AQL: Cookbook
- Legal Notice
-
The information provided herein is »AS IS«. The information contained in this document is subject to change without notice. Better, d.o.o. (hereinafter: Better) gives no warranty of any kind with regard to this material, including, but not limited to, the implied warranties of merchantability and fitness for a particular purpose. Better shall not be liable for any errors contained herein, or for incidental or consequential damages in connection with the furnishing, performance or use of this material.
(c) Better - Reproduction, adaptation or translation without a prior written permission is prohibited, except as allowed under the copyright laws.
- Conventions
-
This document uses the following symbolic conventions:
Courier textCourier text indicates filenames and their contents, computer input and output, program code, etc. For example: “To see the status of server, type
ps -ef | grep java.”Italicized text
Italic text indicates document titles, parameters, emphasized text and replaceable text.
bold text
Bold text indicates emphasized text.
<angle brackets>
Angle brackets indicate generic variable names that must be substituted by real values or strings.
- We welcome your comments
-
Your feedback on the information in this guide is important to us. Please send your comments about this guide to make it even clearer and more accurate.
- Browser support
-
Our products work with all the latest major browser releases and do not require any special additions, extensions or previous setup. We discourage using Internet Explorer at all.
We do support:
-
Edge 17+,
-
Chrome 60+,
-
Firefox 56+,
-
Safari 11+,
-
Opera 50+.
-
About
The openEHR community has defined a query language specification based on archetypes called AQL – Archetype Query Language. Records in openEHR systems are based on archetypes, which define the structural and terminological model of the data.
This provides an opportunity to query clinical statements and data values based on content models, independent of applications which produced the data, programming languages, system environment, and storage models.
| Parts of this document are supported only in Better Platform and are not part of core AQL specification. |
Glossary
- Better Platform
-
Better Platform is a big-data, high-performance solution designed to store, manage, query, retrieve and exchange structured electronic health record data based on the latest release of openEHR specifications. All clinical information is stored in vendor-independent archetypes and templates, allowing standard data entry and retrieval with terminology based validation.
See https://platform.better.care - EHR Server
-
EHR Server is the core part of Better Platform, hosting services and exposing the API.
- Sandbox
-
Sandbox is a cloud hosted Better Platform installation. Here you can test most of the API. With a user account you can use and test full Better Platform potential in your own sandbox.
See https://www.better.care/try-sandbox - JSON
-
JavaScript Object Notation - a lightweight format for storing and transporting data
- EHR
- EHR ID
-
Single EHR unique identifier
- Composition
-
Digital document containing clinical data.
- AQL
What is AQL
Archetype Query Language (AQL) is a declarative query language developed specifically for expressing queries used for searching and retrieving the clinical data found in archetype-based EHRs. It is applied to the openEHR Reference Model (RM) and the openEHR clinical archetypes, but the syntax is independent of applications, programming languages, system environment, and storage models.
What can Archetype based querying do?
-
AQL can execute queries against openEHR based clinical data repository.
-
AQL can retrieve data from openEHR based clinical data repository.
-
AQL expresses queries at the archetype level, i.e. semantic level.
-
AQL can utilise containment mechanism to indicate the data hierarchy and constrain the source data to which the query is applied.
-
AQL allows setting query criteria using archetype and node identifiers, data values within the archetypes, and class attributes defined within the openEHR RM.
-
AQL queries are vendor independent and portable across openEHR clinical data repositories.
-
AQL query can return whole Compositions or fine-grained health data elements.
-
AQL can query a single EHR for point of care purposes, or the entire EHR population in the server for health analytics, population health, research or epidemiological studies.
-
AQL enables longitudinal processing of health data, regardless of originating system or application.
AQL as a standard
Although AQL is an openEHR.org specification, there are different flavours of the AQL language.
However, to be compliant with the spec, they all support at least the major syntax (such as SELECT, FROM, CONTAINS, WHERE, ORDER BY) and path syntax to locate nodes or data values within archetypes in a similar manner.
| Most of the AQL implementations also have their own proprietary extensions in addition to the AQL specification! Check the AQL syntax specification for the supported core keywords. |
How querying works
Let’s compare an openEHR based data repository with a standard relational database.
Relational database
In relational database, data is stored in a number of tables spread across the database.
What we can address are data rows (or records), split into separate named columns.
Data rows from different tables can be linked by using foreign keys, where a data row references a different data row by its local id.
This way you can link a number of records to build a complete data document.
A data document may consist of many different pieces of information that constitute a document model (for example, a blood pressure measurement, a body temperature measurement, etc.). Usually each of these substructures are stored in a separate table, with an internal structure/field description than can be mapped to the structure itself.
Whenever you would like to reconstruct such a document, you would need to pull relevant data from multiple tables and bind it together into a single resulting document. A query to achieve that can be pretty complex, with multiple nesting queries and join statements, which may seriously affect the performance.
Each model change in the future could have serious consequences in terms of:
-
modifying the structures of existing data tables,
-
creation of new tables and binding their data to existing data,
-
affecting other data relationships, where internal queries need to be updated,
-
modifying or applying any constraints over new or already existing data,
-
updating restrictions, which could mark existing data as invalid…
Main point to remember is that we can address specific data by its location in the tables. And once the data is stored, any restrictions applies to all data collected the same. Hence, the query depends on physical storage solution instead on the content.
openEHR repository
On the other hand an openEHR based repository is one big bag full of clinical data documents, split down to separate fields/data nodes. These data nodes are grouped together in documents containing a tree-like structure. No tables, no need to know where the data resides.
You do not need to worry HOW and WHERE the data is stored in terms of tables and columns but WHICH clinical concept the data belongs to.
How different parts of document tree are structured depends on the openEHR template used to store that document, but those individual parts are internally structured according to the archetypes.
|
openEHR archetype
A single archetype defines the maximum data set of attributes/fields for a single clinical concept - blood pressure, body temperature, exertion level, diagnosis, etc. Each field is of a specified type and can contain predefined values and restrictions. openEHR template
A template represents the document model. It defines the document structure, data types and possible data restrictions. A template is a collection of fields from different archetypes. When storing an openEHR document, its structure and contents are validated by matching it to a template - after that the validation is no longer applied. Each field in the template can be uniquely addressed within its own archetype. |
When accessing the openEHR data, you can either access full documents or individual data nodes across the whole repository.
AQL
AQL query is very similar in structure to a standard SQL query syntax.
You will recognize the keywords such as SELECT, FROM, WHERE, ORDER BY and others.
The main difference is we do not address the data by its physical location in tables/columns but to its affiliation to specific archetypes and location within them.
AQL paths
The health data record in openEHR system is stored in a form of a composition - think of it as a single logical document committed to an EHR system. A composition conforms to template archetypes, which define the structural and terminological model of the data.
Querying in openEHR is based on archetype structures. Archetype are hierarchical in structure, and every node can be addressed by its path, where the paths are the basis for locating all data.
Every node in an archetype can be referenced by a path.
These paths are not affected by how the archetype is used in templates; queries may be built using archetype paths in such a way that a query will work regardless of what template or templates the archetype might has been used in. This means paths are therefore safe for querying use.
Paths are enabled by presence of node-id on archetype object nodes – the ‘at-codes’.
context/start_time/value content[openEHR-EHR-OBSERVATION.blood_pressure.v2]/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value
AQL query in a nutshell
Queries are expressed using AQL language which is based on SQL and W3C XPath syntax to locate nodes or data values within archetypes.
Like SQL, AQL has SELECT, FROM, WHERE, ORDER BY syntax statement structure:
-
The
SELECTclause specifies the data elements to be returned. -
The
FROMclause specifies the result source and the corresponding containment (CONTAINS) criteria. -
The
WHEREclause specifies data value criteria within the result source. -
The
ORDER BYclause indicates the data items used to order the returned result set.

AQL result set structures
As a result of an AQL you can get different structures - either containers or base elements, representing different data types.
Containers
Containers give the result data tree some internal grouping and structure, while also providing some default attributes - for instance, each POINT_EVENT inherits from EVENT container and has time attribute out of the box.
| Refer to the Reference model - Data structures fork more details. |
Elements
Elements contain data you defined in the template.
Majority of the openEHR data types are primitive, for example DV_TEXT, DV_COUNT, DV_DATE, DV_TIME, DV_BOOLEAN.
Their value can be expressed as a string, integer, date, time or a boolean.
Some elements are complex, such as DV_QUANTITY, DV_PROPORTION and DV_DURATION.
They have different attributes besides value.
DV_QUANTITY has a decimal magnitude, integer precision and string units, which contains a unit description. DV_PROPORTION`has a `numerator and a denominator.
| See openEHR Base Types specification for more details. |
Result set structure depends on what you queried for.
SELECT
c/context/start_time
FROM COMPOSITION c
LIMIT 1[
{
"#0": {
"@class": "DV_DATE_TIME",
"value": "2014-03-02T04:39:37.000+01:00"
}
}
]SELECT
c/context/start_time as creationTime
FROM COMPOSITION c
LIMIT 1[
{
"creationTime": { (1)
"@class": "DV_DATE_TIME",
"value": "2014-03-02T04:39:37.000+01:00"
}
}
]| 1 | Element of the query result set is now named with alias name |
SELECT
c/context/start_time/value as creationTime (1)
FROM COMPOSITION c
LIMIT 1| 1 | We added the /value selector |
[
{
"creationTime": "2014-03-02T04:39:37.000+01:00"
}
]Result is not a node object but plain value.
SELECT
o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as systolic
FROM OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]
LIMIT 1[
{
"systolic": {
"@class": "DV_QUANTITY",
"magnitude": 125.0,
"units": "mm[Hg]",
"precision": 0
}
}
]As you can see, Systolic field is represented by a DV_QUANTITY openEHR data type and here are listed its main attributes.
SELECT
o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as systolic,
o/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value as diastolic,
c/context/start_time/value as creationTime
FROM COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]
LIMIT 1[
{
"systolic": {
"@class": "DV_QUANTITY",
"magnitude": 125.0,
"units": "mm[Hg]",
"precision": 0
},
"diastolic": {
"@class": "DV_QUANTITY",
"magnitude": 85.0,
"units": "mm[Hg]",
"precision": 0
},
"creationTime": "2019-08-02T13:49:57.428616+02:00"
}
]SELECT
o/data[at0001]/events[at0006]/data[at0003] as event
FROM OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]
LIMIT 1[
{
"event": {
"@class": "ITEM_TREE",
"name": {
"@class": "DV_TEXT",
"value": "blood pressure"
},
"archetype_node_id": "at0003",
"items": [
{
"@class": "ELEMENT",
"name": {
"@class": "DV_TEXT",
"value": "Systolic"
},
"archetype_node_id": "at0004",
"value": {
"@class": "DV_QUANTITY",
"magnitude": 125.0,
"units": "mm[Hg]",
"precision": 0
}
},
{
"@class": "ELEMENT",
"name": {
"@class": "DV_TEXT",
"value": "Diastolic"
},
"archetype_node_id": "at0005",
"value": {
"@class": "DV_QUANTITY",
"magnitude": 85.0,
"units": "mm[Hg]",
"precision": 0
}
}
]
}
}
]The event data and underlying structures are defined by the openEHR archetypes, the data points are expressed as openEHR data types.
Access the data
openEHR Reference Model defines object classes that support your work with the clinical data.
| openEHR RM class name | description |
|---|---|
EHR |
The root object and access point of an EHR for a subject of care. See EHR Class. |
COMPOSITION |
Primary ‘data container’ for clinical content. Instances of the COMPOSITION class can be considered as self-standing data entries. See COMPOSITION Class. |
SECTION |
Provides logical structure to organize and navigate entries. See SECTION Class. |
OBSERVATION, EVALUATION, INSTRUCTION, ACTION |
Clinical statement entry type. See class descriptions. |
ADMIN_ENTRY |
Administrative information entry type. See ADMIN_ENTRY Class. |
Simple data access queries:
SELECT c
FROM EHR e
CONTAINS COMPOSITION cList all compositions contents.
The query above assigns a variable name e to EHR class objects. However EHR class objects are not used/referenced anywhere further, therefore we do not need them.
The example bellow is an equivalent to the example above:
SELECT c
FROM COMPOSITION cList all compositions contents.
Similarly, when we query the OBSERVATION object, instead of writing:
SELECT o
FROM EHR e
CONTAINS COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]we could shorthand it to:
SELECT o
FROM OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]We did not need references to EHR and COMPOSITION, therefore we can leave them out.
Other important classes are VERSIONED_OBJECT and VERSION, which allow us to access composition versioning information.
Composition structure
A data document - composition - is a tree-like data structure. It has a top entry and each of the nodes is either a container or an element node. Each of the nodes can be addressed by the path, defined in the archetype that covers that node. This archetype defined path does not change, regardless of how we include the data point in the template.
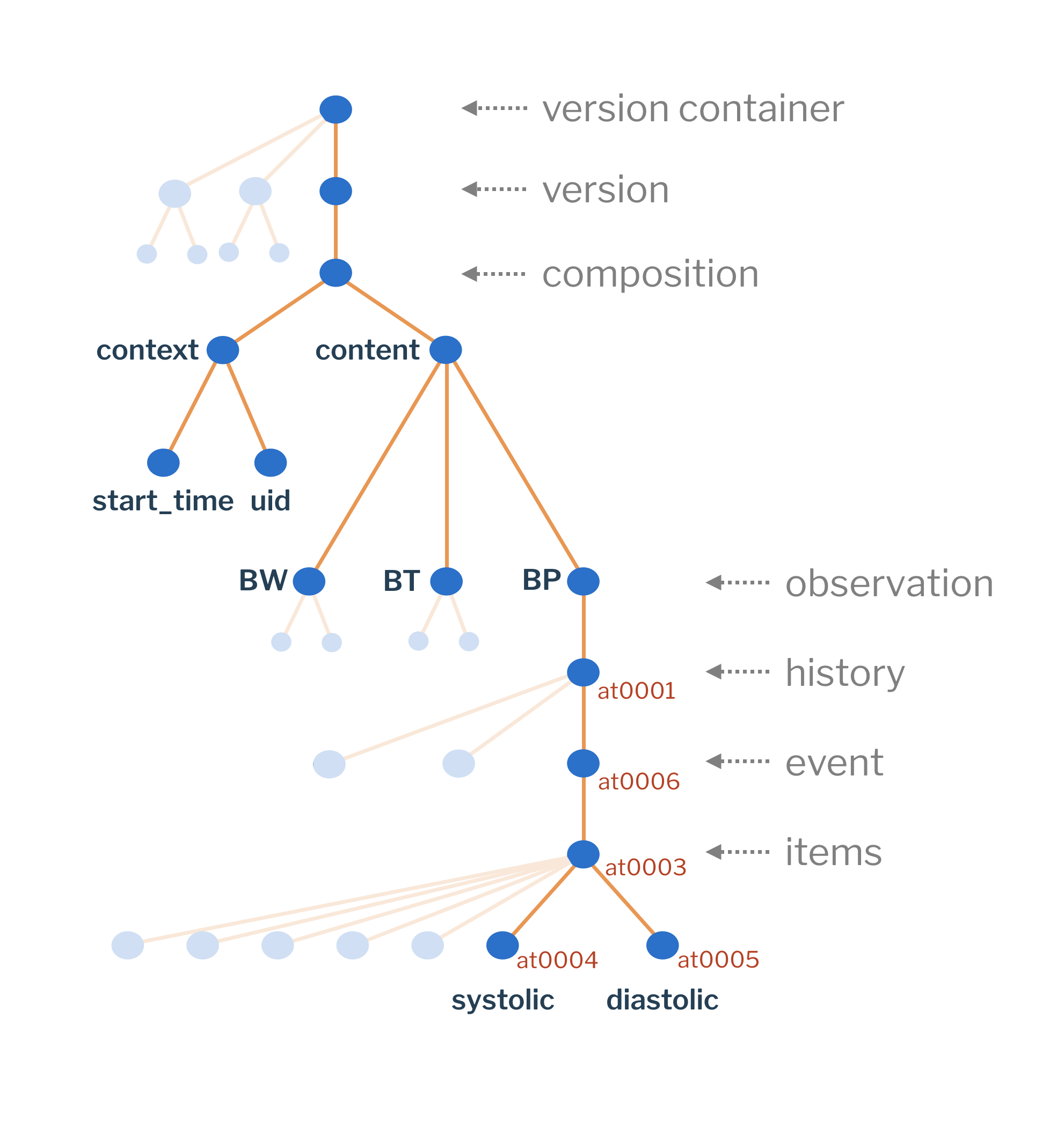
| level | description |
|---|---|
version container |
This node is direct descendant of the EHR and is represented by |
version |
This node represents a single document version and is represented by |
composition |
Composition represents the clinical data part of the single document version, splitting it into the metadata (context) and clinical data (content). |
observation |
This node and underlying data structures are defined by one or many different archetypes in a single or multiple instances of each. |
-
BP - blood pressure observation data node,
-
BT - body temperature observation data node,
-
BW - body weight observation data node.
Their sub-structure is defined by the archetypes and you cannot modify it (the 'at-codes) in a template representing your model. You could rename the nodes, but that would not change the structure itself.
Accessing the data
You can access composition version information, previous versions, its EHR information, context, audit data and the clinical data with references to them - AQL paths.
Some sample paths:
context/start_time/value
context/cid/value
content[openEHR-EHR-OBSERVATION.blood_pressure.v2]/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value
If the composition contains a blood pressure observation, we are accessing the systolic data point following the path expressed by the red 'at-codes', which are defined in the blood pressure archetype.
How is the data stored
As we saw a composition can contain multiple containers and each of them can contain other structures - it all depends on the template.
In following case we are looking at an OBSERVATION based structure (defined by OBSERVATION Class), which allows multiple EVENT (list item) entries in the HISTORY (list container). Other archetypes might have different content.
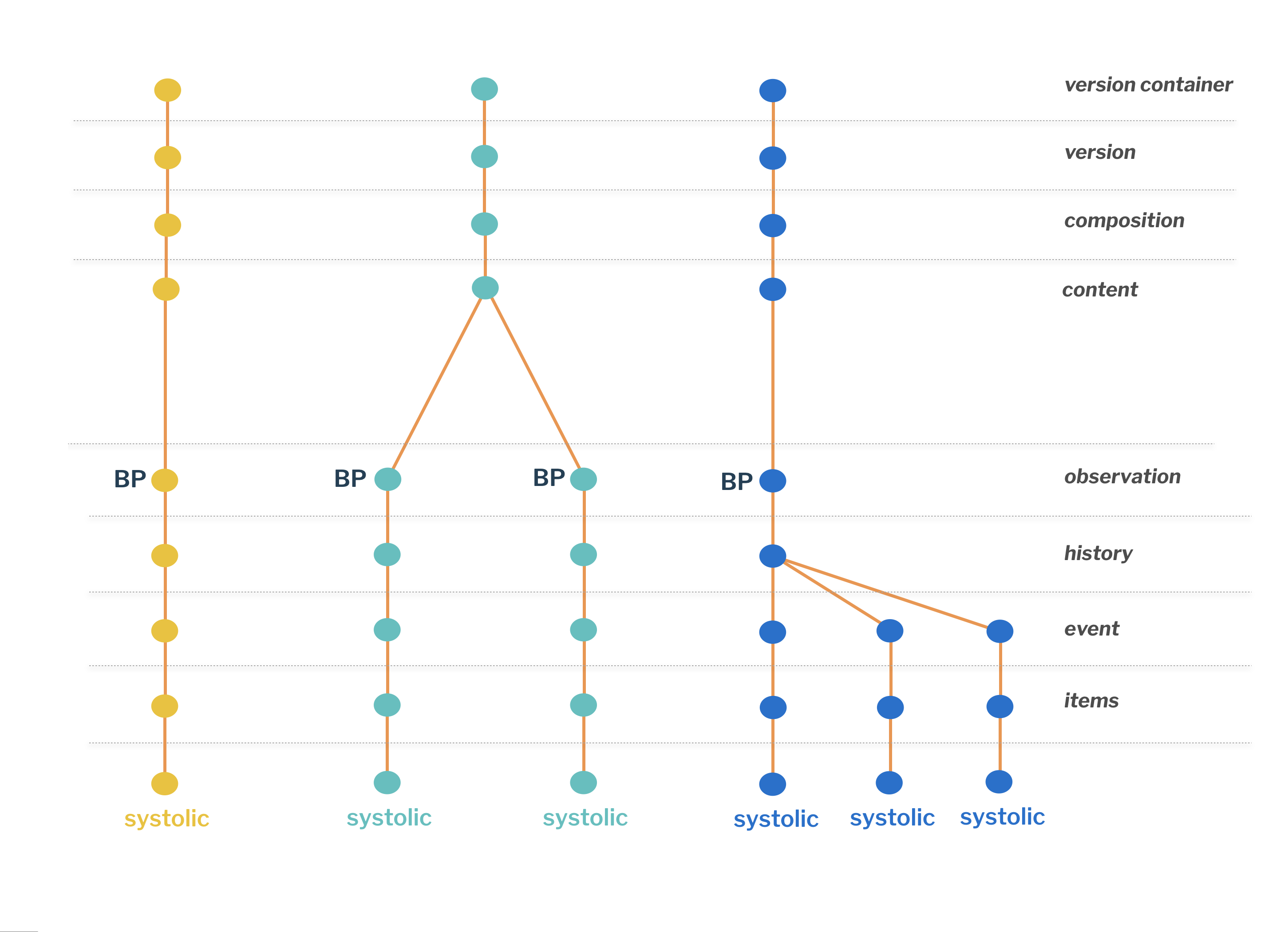
Most common scenario is where a composition contains a single OBSERVATION containing a single EVENT.
A composition can contain multiple measurements of the same type (or different types), each stored in a single OBSERVATION structure. In our sample, each observation contains a single EVENT entry.
| One of the typical situations to use such structure in a document would be different test scenarios, where you need to collect vital signs data before the test and a set after the test. These measurement pairs represent the whole scenario and it would make sense we stored them together. |
A composition could contain a single OBSERVATION which can contain multiple EVENT entries of the same type (in our case it is blood pressure, but could be body temperature, body weight, etc.), each holding a list of data elements - systolic is one of them.
| This would usually be the case when multiple measurements have been taken in a short period of time but were not recorded individually. |
Retrieving data from sub-structures
All in all we stored six separate systolic measurements in three documents and a single query can access them all:
SELECT o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as systolic
FROM COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]Reference entry point
The composition is the primary data container for clinical content.
The key information in a composition is found in its content and context attributes.
Compare the following two queries:
SELECT c/content[openEHR-EHR-OBSERVATION.blood_pressure.v2]/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value
FROM COMPOSITION cWe have addressed a systolic field starting from the composition content part, filtering it to the openEHR-EHR-OBSERVATION.blood_pressure.v2 with a predicate.
SELECT o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value
FROM COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]We have filtered the openEHR-EHR-OBSERVATION.blood_pressure.v2 composition content part with CONTAINS clause, named that node o and referenced it in the SELECT statement.
Both queries return the same data!
| attribute name | description |
|---|---|
|
Composition’s instance unique identifier. |
|
Composition’s name, often same as |
|
Mandatory indicator of the localized language in which this composition is written. |
|
Name of territory in which this Composition was written. ISO 3166 standard |
|
Indicates what broad category this composition belongs to, e.g. persistent - of longitudinal validity, event, process, … |
|
Context information of a healthcare event involving the subject of care and the health system. |
|
Contains relevant template archetype identification information. |
|
Globally unique archetype identifier to which composition complies |
|
Globally unique template identifier. |
|
Start time of the clinical session or other kind of event during which a provider performs a service. |
|
The person responsible for the content of the composition. |
AQL syntax
Case sensitivity
AQL keywords are not case sensitive. All following statements are valid:
SELECT e FROM EHR e ORDER BY e/time_created/value select e from ehr e order by e/time_created/value sELecT e from EHR e ORdeR bY e/time_created/value
Aliases and AQL paths are case sensitive.
SELECT
o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as Systolic
FROM OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]
ORDER BY Systolic/magnitudeSELECT
O/data[at0001]/events[at0006]/DATA[at0003]/items[at0004]/value as Systolic (2)
FROM OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2] (1)
ORDER BY SYSTOLIC/magnitude (3)| 1 | OBSERVATION alias is declared as lower-case o. |
| 2 | Upper-case alias O is not declared.
Also AQL path part /DATA[at0003] should be data[at0003] as specified in the archetype. |
| 3 | Invalid SYSTOLIC alias reference, since the declared one is Systolic. |
Comments
Use standard SQL comments - anything following the double dash `--`is ignored.
SELECT
o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as Systolic -- take only a single field
FROM OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2] -- focus on blood pressure
ORDER BY Systolic/magnitude -- do not forget, you cannot sort on complex typesSimple query
SELECT c
FROM COMPOSITION cThe FROM clause specifies the data source.
The example assigns a variable name c to COMPOSITION class objects. Variable c is used by the SELECT clause as to retrieve composition objects.
This statement would translate to:
Load all compositions from the repository.
You can add more constraints and criteria to the query.
Query criteria
Query criteria can be expressed in two ways:
-
as a
WHEREclause, -
as a class based predicate.
WHERE clause and operators
SELECT c
FROM EHR e
CONTAINS COMPOSITION c
WHERE e/ehr_id/value = "e119f88b-36b7-4537-9914-22bb9396e101"List all compositions belonging to a specific EHR
The WHERE clause syntax has the following parts (in order): keyword WHERE and identified expression(s).
Boolean operators (AND, OR, NOT) and parenthesis can be used to represent multiple identified expressions.
Operators
When setting up the WHERE clause we can combine multiple conditions.
AND operatorSELECT c/uid/value as compositionId,
c/context/start_time/value as creationTime
FROM EHR e
CONTAINS COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v2]
WHERE creationTime >= '2013-08-11T22:24:35.000'
AND
creationTime <= '2014-01-01'List all document IDs for compositions containing body temperature OBSERVATION from the specified time frame.
SELECT
c/context/start_time/value as creationTime,
o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as temperature
FROM EHR e
CONTAINS COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v2]
WHERE (
creationTime >= '2013-08-11T22:24:35.000'
AND
creationTime <= '2014-01-01'
)
AND
(
temperature/magnitude < 36
OR
temperature/magnitude > 40
)List all entries with temperatures less than 36 or above 40, from the specified time frame.
EXISTS operatorThere are no null values in the composition.
The node with the expected data simply does not exist.
Therefore we do not check with a != operator but we use the EXISTS operator.
SELECT
o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as temperature,
o/protocol[at0020]/items[at0021]/value as siteOfMeasurement
FROM EHR e
CONTAINS COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v1]
WHERE EXISTS siteOfMeasurementList only entries that do have the siteOfMeasurement data.
Do not use the condition WHERE siteOfMeasurement != ""
|
NOT operatorWe could use the above query and substitute the WHERE clause with
...
WHERE NOT EXISTS siteOfMeasurementList only entries that do not include the siteOfMesurement data.
Following two queries are equal:
!= operatorSELECT DISTINCT c/archetype_details/template_id/value as templateName
FROM EHR e
CONTAINS COMPOSITION c
WHERE templateName != "Vital Signs"List only template names used that do not match "Vital Signs".
NOT operatorSELECT DISTINCT a/archetype_details/template_id/value as templateName
FROM EHR e
CONTAINS COMPOSITION a
WHERE NOT (templateName = "Vital Signs")List only template names used that do not match "Vital Signs".
Negation operator NOT has precedence over comparison, therefore wrap the condition into parenthesis!
|
LIKE operatorAllows matching with wildcards * and ?:
-
*stands for any string, -
?stands for any character.
SELECT DISTINCT c/archetype_details/template_id/value as templateName
FROM EHR e
CONTAINS COMPOSITION c
WHERE templateName LIKE "Vital*"List all template names used that start with "Vital" substring.
Class based predicate
SELECT c
FROM EHR e[ehr_id/value = 'e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION cList compositions belonging to a specific EHR ID.
SELECT c
FROM EHR e
CONTAINS COMPOSITION c
WHERE e/ehr_id/value = "e119f88b-36b7-4537-9914-22bb9396e101"List compositions belonging to a specific EHR ID.
Both queries are equivalent.
Clause predicate can be used on EHR level and on CARE_ENTRY level.
Predicates on the COMPOSITION level are not supported.
Parsing and executing predicates on class level instead with the WHERE clause can impact the performance!
|
When addressing an observation, we must set its archetype type predicament.
SELECT o
FROM COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]List all blood pressure data.
OBSERVATION Class object defines two attributes, context and content and we can address them directly.
But beware, this way we are referencing the specific observation location within the composition!
SELECT c/content[openEHR-EHR-OBSERVATION.blood_pressure.v2]
FROM COMPOSITION cList all blood pressure data that is located directly under the
contentnode.
The latter example expects blood pressure observations located directly on the content node.
If your template has other content there (i.e. sections), this query will not return any results.
Use generic queries to get data ignoring its location within the composition.
|
Sorting
Whenever you execute a query, the results are returned in a random fashion. If you do not order the results in some way, there is no way to know about their sequence order.
Use ORDER BY clause to sort the results and make sure about the order of the result set items.
The keyword ORDER BY is followed by an identified path and the keyword:
-
ASCorASCENDINGfor ascending order, -
DESCorDESCENDINGfor descending order.
SELECT c
FROM EHR e[ehr_id/value = 'e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION c
ORDER BY c/context/start_time/value DESCList compositions belonging to a specific EHR ID and order it by time of creation, descending
| When trying to sort by complex types (see Elements), the response might be an error or unsorted result set. In case of complex types always refer to the type primitive components (strings, numbers, …). |
Aliases
Use aliases to name result set parts.
SELECT c/context/start_time/value AS creationTime,
e/ehr_id/value AS ehrId
FROM EHR e
CONTAINS COMPOSITION cList all compositions creation time and EHR ID they belong to.
You can also use aliases in WHERE and ORDER BY clauses.
SELECT c/context/start_time/value AS creationTime,
e/ehr_id/value AS ehrId
FROM EHR e
CONTAINS COMPOSITION c
WHERE ehrId = "e119f88b-36b7-4537-9914-22bb9396e101"
ORDER BY creationTime DESCList all compositions creation time and EHR ID for a specific EHR ID, sort them by time of creation, descending.
Limiting the output
In order to limit the number of result set entries you can use different clauses.
TOP clause
Use immediately after the SELECT keyword and pass the number of desired entries.
SELECT TOP 5 c/context/start_time/value as creationTime,
c/uid/value as compositionId
FROM COMPOSITION c
ORDER BY creationTime DESCList latest 5 compositions creation time and composition ID.
Use FORWARD or BACKWARD parameter combined with TOP to select from the top or from the bottom of the list.
If none is used, the default is FORWARD.
SELECT TOP 5 BACKWARD c/context/start_time/value as creationTime,
c/uid/value as compositionId
FROM COMPOSITION c
ORDER BY creationTime DESCList oldest 5 compositions creation time and composition ID
FETCH / LIMIT, paging with OFFSET
Use any of the two terms at the end of the AQL query.
SELECT c/context/start_time/value as creationTime,
c/uid/value as compositionId
FROM COMPOSITION c
ORDER BY creationTime DESC
LIMIT 5List latest 5 compositions creation time and composition ID.
In order to support paging, use the OFFSET clause together with FETCH / LIMIT.
SELECT c/context/start_time/value as creationTime,
c/uid/value as compositionId
FROM COMPOSITION c
ORDER BY creationTime DESC
OFFSET 10 LIMIT 5List 5 compositions creation time and ID, starting with tenth.
Example queries
EHR queries
List patients EHR IDs
SELECT e/ehr_id/value
FROM EHR eList all EHR IDs.
The EHR object is the root object and access point of an EHR for a subject of care (i.e. patient).
The EHR object records three pieces of information that are immutable after creation:
-
system_id- the identifier of the system (domain) in which the EHR was created, -
ehr_id- the identifier of the EHR (distinct from any identifier for the subject of care), and -
time_created- the time of creation of the EHR
| See EHR Information Model for more details. |
One of the basic principles of openEHR is the complete separation of EHR and demographic information, such that an EHR taken in isolation contains little or no clue as to the identity of the patient it belongs to.
It is a common practice to assign a globally unique identifier (GUID) as patients EHR identifier ehr_id/value.
Find patient’s EHR ID and external subject identifier
SELECT
e/ehr_id/value,
e/ehr_status/subject/external_ref/id/value
FROM EHR eList all EHR IDs and their external references.
openEHR EHR does not store patient demographic information (such as name, address, etc). Instead, it supports 2 ways of working with patients:
-
Store patient ID from an external demographic or identity service into the EHR subject field
e/ehr_status/subject, -
Store EHR ID into external demographic server.
External reference as subjectId can be passed at the time of EHR creation and we strongly encourage you to do so.
Count patients with compositions
SELECT COUNT(DISTINCT e/ehr_id/value)
FROM EHR e
CONTAINS COMPOSITION cCount patients that have at least one composition.
Count patients with compositions based on a specific template
SELECT COUNT(DISTINCT e/ehr_id/value)
FROM EHR e
CONTAINS COMPOSITION c[openEHR-EHR-COMPOSITION.encounter.v1]
WHERE
c/name/value = 'Vital Signs'Count patients that have at least one composition based on template "Vital Signs".
Count patients with specific data
SELECT COUNT(DISTINCT e/ehr_id/value)
FROM EHR e
CONTAINS COMPOSITION c
CONTAINS OBSERVATION t[openEHR-EHR-OBSERVATION.body_temperature.v1]Count patients that have at least one composition containing body temperature observation.
List data for multiple patients
As an alternative to predicate constraint we can use MATCHES operator in WHERE statement to constrain an identified path (i.e. ehr_id/value) to match specific values.
The example bellow is equivalent to the example above but allows for multiple EHR IDs:
SELECT e/ehr_status/subject/external_ref/id/value
FROM EHR e
WHERE e/ehr_id/value MATCHES {"e119f88b-36b7-4537-9914-22bb9396e101"}List the external reference for a specific EHR.
SELECT e/ehr_status/subject/external_ref/id/value
FROM EHR e
WHERE e/ehr_id/value MATCHES {'<ehrId1>, <ehrId2>, <ehrId3>'}List the external references for a specific EHRs.
COMPOSITION queries
Count the number of compositions for a specific patient
SELECT COUNT(c)
FROM EHR e[ehr_id/value='e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION cCount all compositions for a specific EHR.
The FROM clause assigns a variable name e to EHR class objects and uses a predicate to constrain ehr_id/value to a chosen value.
A variable name c is assigned to COMPOSITION class objects.
A COUNT() function in SELECT clause is used to return the number of COMPOSITION class objects from the result set.
List all compositions for a specific patient
SELECT c
FROM EHR e[ehr_id/value='e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION cList all compositions for a specific EHR.
Within EHR class you need to determine whether this query is applied to a single EHR or all EHRs.
The latter is also called a population query.
If it is for all EHRs, you don’t need to specify ehr_id/value in the FROM clause.
In this example we query for a single patient EHR and we use a predicate to constrain ehr_id/value to match the selected patient.
It is a common practice to assign a globally unique identifier (GUID) as patient EHR identifier in ehr_id/value.
List five latest compositions for a specific patient
SELECT TOP 5 c
FROM EHR e[ehr_id/value='e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION c
ORDER BY c/context/start_time DESCList 5 latest compositions for a specific EHR.
The SELECT TOP statement is used to limit the number of items from the result set.
It starts with keyword TOP, followed by an integer number and/or the direction (i.e. BACKWARD, FORWARD).
The FROM clause uses a predicate to constrain ehr_id/value to specific patient’s EHR ID.
We assigned a variable name c to instances of COMPOSITION which are ordered by clinical session date stored in context/start_time attribute in descending order.
List composition from a specified time frame
SELECT c
FROM EHR e
CONTAINS COMPOSITION c
WHERE
c/context/start_time >= '2013-08-11T22:24:35.000' AND
c/context/start_time <= '2014-01-01'List all compositions created between two dates.
The WHERE clause is used to set date-time range criteria for clinical session which are recorded in composition’s context/start_time attribute.
Date-time literals are enclosed in single quotation marks and formatted as ISO date/time string.
List all unique templates used for the compositions
SELECT DISTINCT c/archetype_details/template_id/value
FROM COMPOSITION cList all used template names.
A data composition conforms to a template - which represents a structural and terminological model for a specific type of a clinical record. A template is used to logically represent a use case specific data set, such as the data items making up a record for Discharge Summary, Vital Signs, Laboratory Report and others.
Templates have globally unique template id and are based on archetypes - constructed by referencing relevant items from a number of archetypes.
Compositions carry metadata relevant to the structure in the archetype_details attribute.
The template_id holds the template unique identifier.
The SELECT clause references a path within a composition where template identifier is stored.
In addition, the DISTINCT clause is used to remove duplicates from the result set.
List compositions based on specific template for a patient
SELECT c
FROM EHR e[ehr_id/value='e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION c[openEHR-EHR-COMPOSITION.encounter.v1]
WHERE c/name/value='Vital Signs'List compositions that belong to a specific patient and the template used to validate it was Vital Signs.
The CONTAINS clause specifies the data source and the corresponding containment criteria to specify the data source from which the required data is to be retrieved.
It assigns a variable name c to COMPOSITION class objects and uses an archetype predicate to constrain query to compositions based on a specific archetype - openEHR-EHR-COMPOSITION.encounter.v1.
There might be many templates based on the same archetype, therefore additional WHERE clause with identified path is used to specify required title - 'Vital Signs'.
List compositions containing specific observations for a specific patient
SELECT c
FROM EHR e[ehr_id/value='e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v2]List all compositions that contain body temperature data.
The example utilizes a nested containment constraint to specify the hierarchical relationships between parent and child data items.
The FROM clause assigns a variable name e to EHR class objects and uses a predicate to constrain ehr_id/value to a chosen patient.
The first CONTAINS clause assigns a variable name c to COMPOSITION class objects which must satisfy containment criteria from the second CONTAINS clause.
The second CONTAINS clause assigns a variable name o to OBSERVATION class objects and uses an archetype predicate to constrain further to compositions with data items matching requested archetype - openEHR-EHR-OBSERVATION.body_temperature.v2.
List patients with compositions containing multiple observations
SELECT ehr_id/value
FROM EHR e
CONTAINS COMPOSITION c
CONTAINS (
OBSERVATION t[openEHR-EHR-OBSERVATION.body_temperature.v2]
AND
OBSERVATION bp[openEHR-EHR-OBSERVATION.blood_pressure.v2]
)List all patients that have compositions containing BOTH body temperature and blood pressure data.
The containment constraint is constructed using a keyword CONTAINS between two class expressions. Left class expression is the parent object of the right class expression. Boolean operators (AND, OR, NOT) and parentheses are used when multiple containment constrains are required.
The FROM clause assigns a variable name e to EHR class objects. The query is applied to all EHRs (population query) and returns the ones containing compositions with data items matching archetype for body temperature (i.e openEHR-EHR-OBSERVATION.body_temperature.v2) and blood pressure (i.e openEHR-EHR-OBSERVATION.blood_pressure.v2).
List patients with body temperature greater than 39
SELECT e/ehr_id/value
FROM EHR e
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v2]
WHERE o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value/magnitude > 39List all patients with temperature higher than 39
A WHERE clause is used to represent criteria applied to the data items.
Variable o is assigned the OBSERVATION class object.
An identified path o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value/magnitude which points to body temperature magnitude is used to locate data item within archetype openEHR-EHR-OBSERVATION.body_temperature.2 and require it to be greater than 39.
Body temperature field is of DV_QUANTITY type, which means that its value will be a composite of all its components string representation (magnitude and units).
Address specific component for the correct criteria definition.See Elements for more details! |
|
Query readability
Informative comments within AQL path predicates can be used to increase readability.
AQL path predicates enriched with informative comment using a pair of
Content enclosed by a pair of |
Select patients with abnormal values
SELECT
bp/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value AS systolic,
bp/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value AS diastolic
FROM EHR e
CONTAINS OBSERVATION bp[openEHR-EHR-OBSERVATION.blood_pressure.v2]
WHERE
systolic/magnitude >= 140 AND
diastolic/magnitude >= 90List patients with abnormal blood pressure values systolic pressure >= 140 and diastolic pressure>= 90.
List patients matching a specific problem diagnosis code
SELECT e/ehr_id/e
FROM EHR e
CONTAINS EVALUATION d[openEHR-EHR-EVALUATION.problem_diagnosis.v1]
WHERE
d/data[at0001]/items[at0002]/value/defining_code/code_string MATCHES {'I10', 'E11.9', 'R07.1'}List patients with diagnosis code matching hypertension (I10), Type 2 diabetes mellitus (E11.9) or Chest pain on breathing (R07.1) .
Make sure the template in question linked the Diagnosis name field to an ICD10 terminology which provides the codes in question.
If the field is used as a free text field, the condition might read:WHERE d/data[at0001]/items[at0002]/value matches {'Hypertension', 'Diabetes - type 2', 'Chest pain'}See Terminology query for more specific terminology related information. |
Multiple compositions in a single result set
Obtaining data from different observations in a single document is simple enough.
How to join data from separate compositions?
SELECT e/ehr_id/value, alg, med
FROM EHR e
CONTAINS
(COMPOSITION c1
CONTAINS EVALUATION alg[openEHR-EHR-EVALUATION.adverse_reaction-allergy.v1]
AND
COMPOSITION c2
CONTAINS INSTRUCTION med[openEHR-EHR-INSTRUCTION.medication.v1]
)List of patient’s allergies as well as medications
This type of query has an EHR at the top of the containment and two containment branches joined by a logical AND.
Since c1 and c2 are actually not used, you can write this query as:
SELECT e/ehr_id/value, alg, med
FROM EHR e
CONTAINS
(EVALUATION alg[openEHR-EHR-EVALUATION.adverse_reaction-allergy.v1]
AND
INSTRUCTION med[openEHR-EHR-INSTRUCTION.medication.v1]
)The result of this query will be a cartesian product of all available alg and all available med for each patient. To avoid this, specify additional criteria to limit their number.
An example of a query, where such a limit comes handy is finding the latest blood pressure measurements for patients with a diagnosis of hypertension:
SELECT
e/ehr_id/value,
diag/data[at0001]/items[at0002]/value as diagnosis,
bp/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as systolic,
bp/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value as diastolic
FROM EHR e[ehr_id/value='e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS
(TOP 1 COMPOSITION c1
CONTAINS EVALUATION diag[openEHR-EHR-EVALUATION.problem_diagnosis.v1]
AND
TOP 1 COMPOSITION c2
CONTAINS OBSERVATION bp[openEHR-EHR-OBSERVATION.blood_pressure.v2]
)
WHERE diag/data[at0001]/items[at0002]/value/defining_code/code_string like 'I10*'
ORDER BY c1/context/start_time DESC,
c2/context/start_time DESCCombine latest blood pressure and specified diagnosis of a specific patient from different compositions in a single entry.
Partial archetype name match
When querying for data based on archetype names, you can use the wildcard *.
In case you stored clinical data into the repository using different versions of same archetype, you started with this query:
v1SELECT
bp/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as systolic,
bp/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value as diastolic
FROM EHR e[ehr_id/value='e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION c
CONTAINS OBSERVATION bp[openEHR-EHR-OBSERVATION.blood_pressure.v1]
LIMIT 5List 5 entries for specific patient systolic and diastolic data, using archetype version
v1.
After the template update you stored data using the archetype v2 and you also queried the data using the new archetype name.
If you wanted to access the data stored with previous version and the new one, the query would look like this:
SELECT
bp1/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as systolic1,
bp1/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value as diastolic1,
bp2/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as systolic2,
bp2/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value as diastolic2
FROM
EHR e[ehr_id/value='e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION c
CONTAINS (
OBSERVATION bp1[openEHR-EHR-OBSERVATION.blood_pressure.v1]
OR
OBSERVATION bp2[openEHR-EHR-OBSERVATION.blood_pressure.v2]
)
LIMIT 5List 5 entries for specific patient systolic and diastolic data, using archetype version
v1and archetypev2but use separate variable for each version.
You got the data but you got it in separate variables, which means you need to adapt your application to read data like this and merge it somehow.
Instead, to avoid changing the app, handle such change on query level and produce single variable for a field, across two different archetype versions!
SELECT
bp/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value as systolic,
bp/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value as diastolic
FROM
EHR e[ehr_id/value='e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION c
CONTAINS OBSERVATION bp[openEHR-EHR-OBSERVATION.blood_pressure.v*] (1)
LIMIT 5List 5 entries for specific patient systolic and diastolic data, using any blood pressure archetype version.
| 1 | A wildcard * is used to cover both v1 and v2 variants of archetype name. |
| Whenever an archetype is updated, the majority of the fields remain where they are, if possible. Therefore we expect systolic and diastolic to remain on the same path. |
|
Archetype name is constructed by three identifiers, separated by a dot
|
VERSION queries
Compositions are immutable but they can can be updated by creating a new composition version containing the latest data. Updated versions are not active anymore, which means that the data they contain will not show up in the queries.
How versioning works
Let’s say our example composition ID is ec86e50e-b708-4cc9-961e-57c9c4fa77a4::andrazk-demo.sandbox.com::1 and it contains information about the body temperature of "73.2 °C".
We realise the mistake when we query the data:
SELECT
o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as temp
FROM COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v2]
LIMIT 1List a single body temperature value.
We would receive a single result of "73.2 °C".
We update the composition with the correct value of "37.2 °C" and the composition ID is now ec86e50e-b708-4cc9-961e-57c9c4fa77a4::andrazk-demo.sandbox.com::2.
| Only the latest version of a composition can be updated! |
We repeat the query and the only result is now "37.2 °C". Previous composition was marked as not active and we do not receive its data as part of the query result set.
If you query a specific version of the composition the data can still be retrieved.
SELECT
o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as temp
FROM COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v2]
WHERE c/uid/value = 'ec86e50e-b708-4cc9-961e-57c9c4fa77a4::andrazk-demo.sandbox.com::1'List body temperature values stored in a specific composition.
The result set will contain complete composition data, even if the composition is not active any more.
Versioning data
The hierarchy and inclusion of the objects is displayed in the Composition structure diagram.
Each composition has a VERSIONED_OBJECT data object attached, which contains all the version related meta-data.
| attribute | description |
|---|---|
|
Composition identifier, unique within the repository. All versions of this composition have the same identifier. |
|
The EHR ID to which this composition belongs to. |
|
The |
VERSIONED_OBJECT includes one or more VERSION objects, which each contain a composition version and meta-data about it.
| attribute | description |
|---|---|
|
Composition version ID. |
|
Reference to the previous version of the composition. |
|
Composition version ID. |
|
Composition version ID. |
|
Composition version creation time. |
Access composition information

To access all the active compositions data, execute the following query:
SELECT
vo/uid/value,
vo/owner_id/id/value,
vo/time_created,
v/uid/value,
v/preceding_version_uid/value,
v/commit_audit/committer,
v/commit_audit/time_committed
FROM EHR e
CONTAINS VERSIONED_OBJECT vo
CONTAINS VERSION vList some VERSIONED_OBJECT and VERSION properties for all active compositions.
By default every query returns data for active compositions only.
SELECT
vo/uid/value as identifier,
v/uid/value as versionId,
v/preceding_version_uid/value as previousVersionId
FROM EHR e
CONTAINS VERSIONED_OBJECT vo
CONTAINS VERSION v[all_versions] (1)
ORDER BY identifier ASC,
v/commit_audit/time_created/value DESCList some VERSION_OBJECT and VERSION properties for all compositions, including inactive one.
| 1 | Make sure to include VERSION all_versions predicate to retrieve non active entries too! |
Data aggregation
AQL queries support following aggregate expressions:
-
AVG- getting an average value on numerical values only -
MIN/MAX- getting single minimal and single maximal value on numerical values only -
COUNT- counting size of the result set -
COUNT DISTINCT- counting the number of distinct values
You can combine multiple aggregate expressions in a single query, but cannot not mix aggregate expressions with regular SELECT statement (except when using GROUP BY statement).
When aggregating values on complex types, do not forget to address their primitive part (magnitude in case of a DV_QUANTITY).
|
AVG
Calculate an average value.
SELECT
AVG(o/data[at0001]/events[at0002]/data[at0003]/items[at0004]/value/magnitude)
FROM EHR e[ehr_id/value = 'e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.body_mass_index.v1]List average value for patient body mass index.
MIN and MAX
Find a minimal or maximal value.
SELECT
MAX(o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value/magnitude) as maxSystolic,
MIN(o/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value/magnitude) as minDiastolic
FROM EHR e[ehr_id/value = 'e119f88b-36b7-4537-9914-22bb9396e101']
CONTAINS COMPOSITION c
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]List maximum systolic and minimum diastolic value for a patient.
COUNT
Count the number of occurrences of specific diagnosis.
SELECT COUNT(d)
FROM EHR e
CONTAINS EVALUATION d[openEHR-EHR-EVALUATION.problem_diagnosis.v1]
WHERE
d/data[at0001]/items[at0002]/value/defining_code/code_string = "I10"Count the number of diagnosis recorded, that match hypertension (code I10)
COUNT DISTINCT
Count the number of distinct patients with specific diagnosis.
SELECT COUNT(DISTINCT e/ehr_id/value)
FROM EHR e
CONTAINS EVALUATION d[openEHR-EHR-EVALUATION.problem_diagnosis.v1]
WHERE
d/data[at0001]/items[at0002]/value/defining_code/code_string = "I10"Count the number of distinct patients with diagnosis recorded as hypertension (code I10).
DISTINCT
List the number of distinct patients with specific diagnosis.
SELECT DISTINCT e/ehr_id/value
FROM EHR e
CONTAINS EVALUATION d[openEHR-EHR-EVALUATION.problem_diagnosis.v1]
WHERE
d/data[at0001]/items[at0002]/value/defining_code/code_string = "I10"List distinct patients with diagnosis recorded as hypertension (code I10).
Data grouping
AQL queries can be extended with EHR Server functions, to support following data grouping expressions:
-
SQUASH- format a cartesian product into grouping by specific value, -
UNION ALL- combine data form multiple queries, -
GROUP BY- get all results containing a certain value also grouped by this value.
SQUASH
When your data model has arrays nested within the arrays, the result set is a cartesian product.
Usually you group values by a specific key on the client side, but SQUASH allows you to group it by the key within the result set itself.
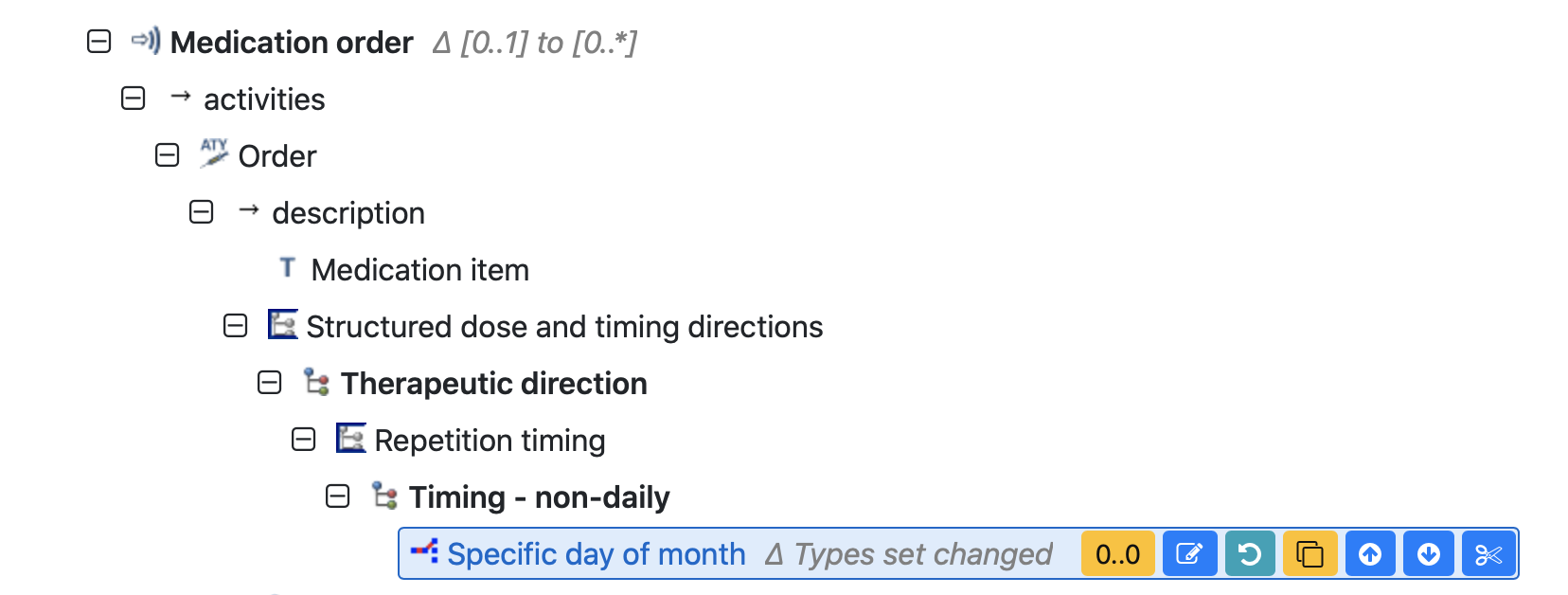
In our case we have a list of Medication orders and each of them can contain a list of days of month, when the medication needs to be administered.
We see that each Medication order can contain a nesting array of Day of week values.
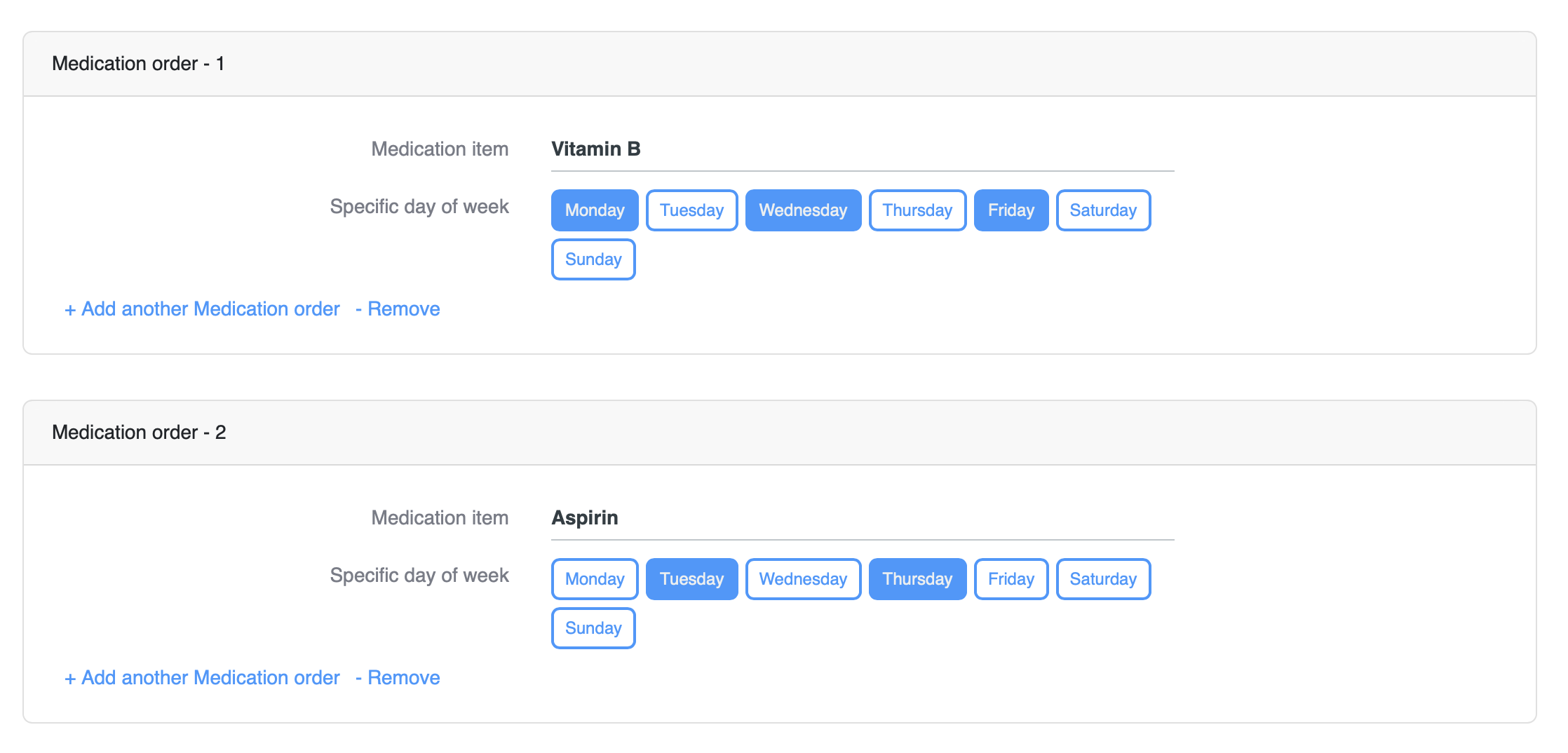
Once the form is filled with the data and composition saved into the repository, we can then execute the query to retrieve the data.
SELECT
ac/description[at0002]/items[at0070]/value/value as medication,
ac/description[at0002]/items[openEHR-EHR-CLUSTER.therapeutic_direction.v1]/items[openEHR-EHR-CLUSTER.timing_nondaily.v1]/items[at0003]/value/value as dayOfWeek
FROM EHR e
CONTAINS COMPOSITION c
CONTAINS INSTRUCTION o[openEHR-EHR-INSTRUCTION.medication_order.v2]
CONTAINS ACTIVITY ac[at0001]List all prescribed medications and their corresponding dayOfWeek values.
The result will be structured like this:
[
{
"medication": "Vitamin B",
"dayOfWeek": "Monday"
},
{
"medication": "Vitamin B",
"dayOfWeek": "Wednesday"
},
{
"medication": "Vitamin B",
"dayOfWeek": "Friday"
},
{
"medication": "Aspirin",
"dayOfWeek": "Tuesday"
},
{
"medication": "Aspirin",
"dayOfWeek": "Thursday"
}
]To pivot data and group it by one of the parent values, use SQUASH function:
SELECT
ac/description[at0002]/items[at0070]/value/value as medication,
SQUASH(ac/description[at0002]/items[openEHR-EHR-CLUSTER.therapeutic_direction.v1]/items[openEHR-EHR-CLUSTER.timing_nondaily.v1]/items[at0003]/value/value) as dayOfWeek
FROM EHR e
CONTAINS COMPOSITION c
CONTAINS INSTRUCTION o[openEHR-EHR-INSTRUCTION.medication_order.v2]
CONTAINS ACTIVITY ac[at0001]List all prescribed medications and their corresponding dayOfWeek values collected in a single array.
[
{
"medication": "Vitamin B",
"dayOfWeek": [
"Monday",
"Wednesday",
"Friday"
]
},
{
"medication": "Aspirin",
"dayOfWeek": [
"Tuesday",
"Thursday"
]
}
]UNION ALL
UNION ALL allows you to combine results from different AQL queries into one.
SELECT
o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value, -- systolic
o/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value -- diastolic
FROM EHR e
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]
WHERE e/ehr_id/value = 'e119f88b-36b7-4537-9914-22bb9396e101'
UNION ALL
SELECT
o/data[at0001]/items[at0002]/value, -- diagnosis
o/data[at0001]/items[at0010]/value -- onsetDate
FROM EHR e
CONTAINS EVALUATION o[openEHR-EHR-EVALUATION.problem_diagnosis.v1]
WHERE e/ehr_id/value = 'e119f88b-36b7-4537-9914-22bb9396e101'List all blood pressure data and diagnosis from different for the same patient. It is similar to Multiple compositions in a single result set joining queries, but we can specify specific criteria for each query separately.
When trying to limit the output of each individual query, use TOP clause instead LIMIT, since latter applies for the combined result set.
|
When using select aliases in UNION ALL query, aliases from the first query will be used in the overall result set.
|
GROUP BY
For simple grouping by a specific field value use the GROUP BY statement.
A couple rules apply:
-
grouping works only on simple values,
-
aggregate values are calculated for each group separately,
-
you can only group by the values that are present in the
SELECTstatement.
| Consult the Better Platform: Developer guide: Grouping result sets for more complex queries grouping requirements. |
SELECT
MIN(o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value/magnitude) as minTemp,
MAX(o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value/magnitude) as maxTemp,
o/protocol[at0020]/items[at0021]/value/value as location
FROM OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v2]
GROUP BY locationList minimal and maximal temperature values grouped by the individual measurement location.
[
{
"minTemp": 36.2,
"maxTemp": 40.1,
"location": "Axilla"
},
{
"minTemp": 36.4,
"maxTemp": 39.3,
"location": "Mouth"
},
{
"minTemp": 35.8,
"maxTemp": 38.7,
"location": "Ear canal"
}
]Text related server functions
For simple text searching related queries check how to use the LIKE operator.
Full text search
Full text search is powered by an internal Lucene engine, that supports advanced search criteria. The purpose of full text search is to support across large text documents that are included in the clinical data - be it in XML, HTML or any other textual format.
Better Platform indexes all fields of type DV_PARSABLE and INSTRUCTION.narrative fields.
| If you search on any other field, that is not of types listed above or one of its descendants, the search will return an empty result set. |
Invoke the search function in the WHERE part of the query, using the following parameters:
FULLTEXT(aqlPath, 'search string')
Search string can contain Lucene supported term modifiers, their default operator is OR.
Let’s say we search across the diagnosis field that contains following values:
viral and other specified intestinal infections (1) viral intestinal infection, unspecified (2) down syndrome, unspecified (3) nonallergic asthma (4)
| search string | description | results |
|---|---|---|
|
Search for any entry that contains a word starting with literal |
1, 2 |
|
Search for any entry that starts with |
4 |
|
Search for any entry that must contain a word |
3 |
|
Search for any entry that contains word starting with |
3 |
|
Search for any entry that contains a word that starts with literal |
2 |
If no other ordering clause is present, the results are ordered by their search score.
For presentation option check the
Better Platform: Developer guide
for the FT_HIGHLIGHT() function.
|
Value concatenation with CONCAT
Server function CONCAT takes a list of arguments and joins them into a string using a single delimiter.
CONCAT(delimiter, arg1, arg2, arg3, ..., argN)
argX can be a literal string or an AQL path to be resolved within the query.
SELECT TOP 1
o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value/magnitude as systolic,
o/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value/magnitude as diastolic,
concat(' ', 'Systolic:', o/data[at0001]/events[at0006]/data[at0003]/items[at0004]/value/magnitude, 'Diastolic:', o/data[at0001]/events[at0006]/data[at0003]/items[at0005]/value/magnitude) as presentation
FROM EHR e
CONTAINS OBSERVATION o[openEHR-EHR-OBSERVATION.blood_pressure.v2]Return a value for systolic, diastolic and presentation string containing both values.
{
"systolic": 120.0,
"diastolic": 80.0,
"presentation": "Systolic: 120.0 Diastolic: 80.0"
}| You can concatenate only primitive types - see Elements. |
Terminology query
In AQLs it is often useful to be able to resolve a set of terminology codes via a terminology REST API call. This way you can include localization or query/filter by custom terminology properties, not just descriptions.
AQL function TERMINOLOGY forwards it’s parameter as a REST API call to the terminology service.
For example, to get all ICD10 descriptions whose code starts with G0, the following AQL can be used:
SELECT c
FROM COMPOSITION c[openEHR-EHR-COMPOSITION.encounter.v1]
CONTAINS EVALUATION e[openEHR-EHR-EVALUATION.problem_diagnosis.v1]
WHERE e/data[at0001]/items[at0002]/value/defining_code/code_string MATCHES TERMINOLOGY('/codesystem/ICD10/entities/query?code=G0&meta.match=start')List all compositions that contain a diagnosis description starting with a description of any code starting with G0