EHR Server API: Cookbook
- Legal Notice
-
The information provided herein is »AS IS«. The information contained in this document is subject to change without notice. Better, d.o.o. (hereinafter: Better) gives no warranty of any kind with regard to this material, including, but not limited to, the implied warranties of merchantability and fitness for a particular purpose. Better shall not be liable for any errors contained herein, or for incidental or consequential damages in connection with the furnishing, performance or use of this material.
(c) Better - Reproduction, adaptation or translation without a prior written permission is prohibited, except as allowed under the copyright laws.
- Conventions
-
This document uses the following symbolic conventions:
Courier textCourier text indicates filenames and their contents, computer input and output, program code, etc. For example: “To see the status of server, type
ps -ef | grep java.”Italicized text
Italic text indicates document titles, parameters, emphasized text and replaceable text.
bold text
Bold text indicates emphasized text.
<angle brackets>
Angle brackets indicate generic variable names that must be substituted by real values or strings.
- We welcome your comments
-
Your feedback on the information in this guide is important to us. Please send your comments about this guide to make it even clearer and more accurate.
- Browser support
-
Our products work with all the latest major browser releases and do not require any special additions, extensions or previous setup. We discourage using Internet Explorer at all.
We do support:
-
Edge 17+,
-
Chrome 60+,
-
Firefox 56+,
-
Safari 11+,
-
Opera 50+.
-
About
Main part of the Better Platform is the EHR Server. It is the core part of the clinical data repository, that incorporates all main services and interfaces:
-
data storage and retrieval,
-
indexing and search features,
-
demographics,
-
security features and access control.
Part of the Better Platform but installed separately are also:
-
Resource Store,
-
Terminology Server,
-
Clinical Decision Support.
All together are accessible via API sets, that are explained in this document. We’ll try to practically demonstrate how to use them and guide you through different situations and explain, how to use which part of the Better Platform to achieve the desired result.
Glossary
- platform, repository
-
Better Platform
- API
-
EHR Server integration REST API
- EHR
- EHR ID
-
Single EHR unique identifier
- CDR
-
Clinical Data Repository - a medical database system designed to provide a realtime summary of a patient’s condition
- XML
-
Extensible Markup Language - a markup language that defines a set of rules for encoding documents in a format that is both human-readable and machine-readable
- JSON
-
JavaScript Object Notation - a lightweight format for storing and transporting data
- SOAP
-
Simple Object Access Protocol - XML-based messaging protocol for exchanging information among computers
- Postman
-
Client side API testing and developing tool - http://www.getpostman.com
- CSV
-
Comma separated value - a file containing delimited text data.
- Reference model
-
Definition of all openEHR classes and data types
How to integrate your app with the Platform?
Imagine a typical GP visit and what kind of interactions happen there between different actors:
-
patient
-
nurse/doctor
-
administrative application/EMR
-
Better Platform, including the Demographics service
Major part of it happens behind the curtains and each specific operation might include multiple steps to be performed.
-
Is the visitor an existing or a new patient?
-
check the demographics entry
-
-
Does this patient already have an EHR?
-
How does a composition look like?
-
How do I correct a mistake in saved data?
-
How do I read the data?
-
Where can I store the images/recordings?
-
How can I manage my terminologies?
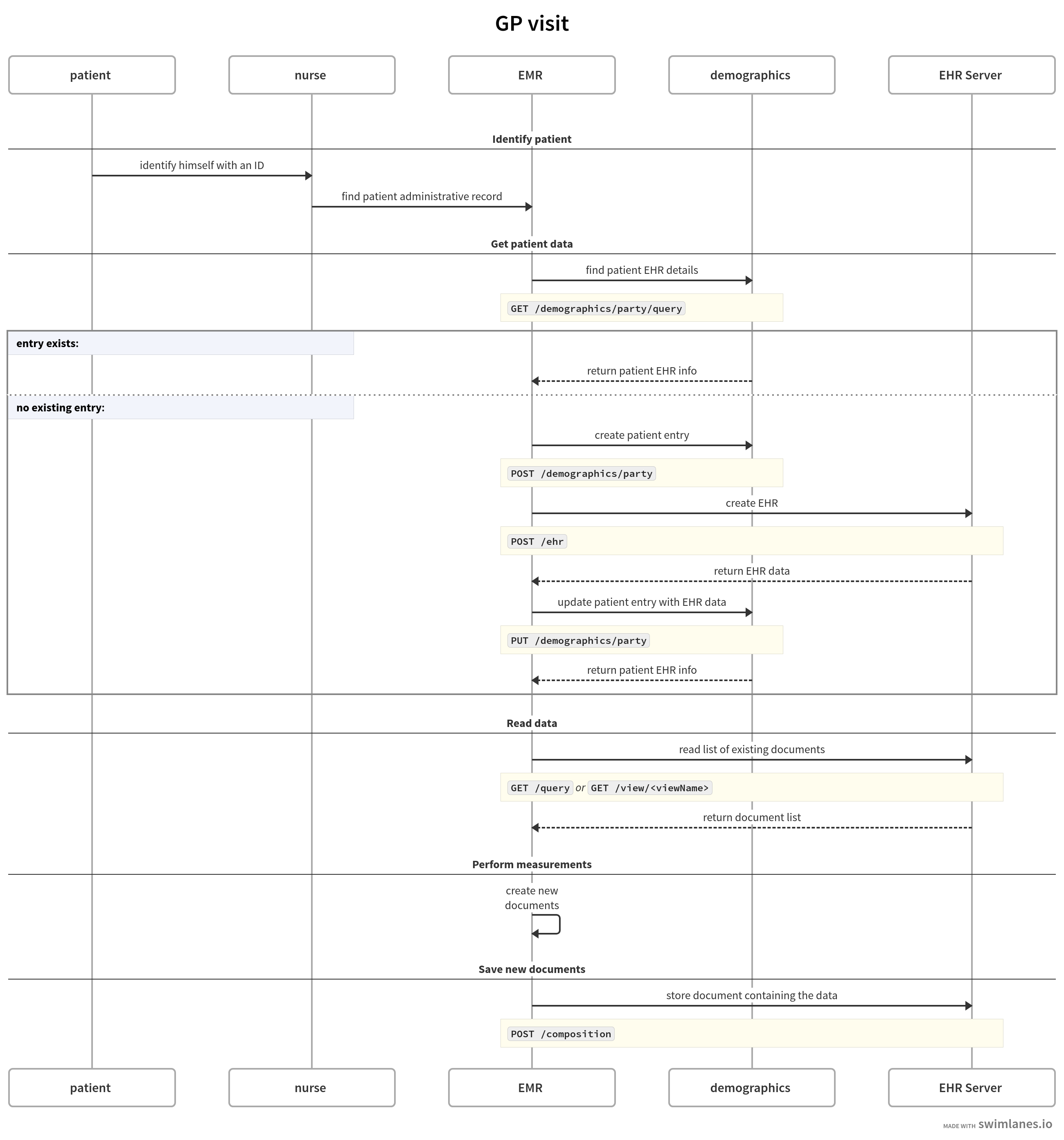
The Platform API sets offer a granular approach to support these operations with simple actions.
Better Platform API sets
In order to interface with the data and services handled by the Better Platform, multiple API sets are available to the developers, depending on the domain you want to cover:
-
Electronic Health Record API to manage patient EHRs, retrieve and store clinical data, execute queries against the CDR and access the demographic data,
-
Terminology API to manage terminology sets and access the data,
-
Resource Store API for storing and retrieving larger binary resources,
-
OpenEHR API implements the standardized openEHR interface.
|
Check Better Platform Sandbox
Go to http://better.care/try-sandbox to register and get access to Better Platform Sandbox environment. |
Electronic Health Record API
EHR API has many endpoints that can facilitate development and data access, but let’s focus on the most used:
-
/ehr to manage patients EHR,
-
/template to access template information, needed for composition structure information,
-
/composition to store and retrieve clinical documents,
-
/query to retrieve any data contained in the CDR,
-
/view to access predefined queries, stored on the EHR Server.
Endpoint /ehr
In physical world, each patient would have a folder somewhere in a drawer cabinet in the doctors office, containing all the clinical history in form of various documents.
OpenEHR repository has an equivalent, the EHR.
Ideally, a single patient should have and be paired with a single EHR. Make sure you do not create multiple EHR for a single patient, because his clinical document would then be spread across multiple containers and thus incomplete in most of the listings.
Let’s say our patient Samantha just visited the doctor and it is her first visit, therefore she has no EHR yet.
Create an EHR
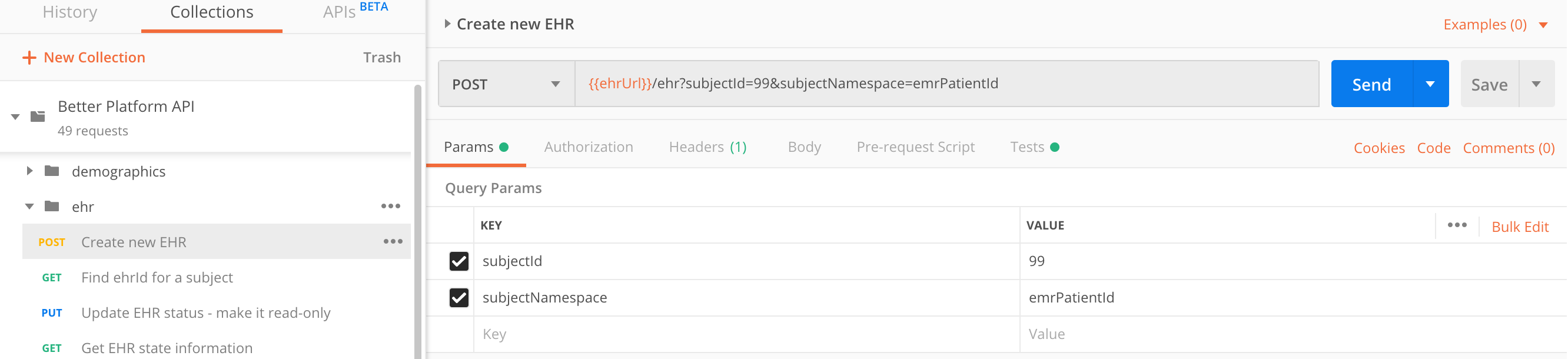
The call to create a new EHR is simple and requires no extra parameters, even though we recommend using subjectId paired with subjectNamespace.
| parameter | type | required | description |
|---|---|---|---|
subjectId |
string |
no |
An unique ID, that is matched with the patient/owner of the EHR in real life. |
subjectNamespace |
string |
no |
Define the domain of the |
committerName |
string |
no |
Store the information about the person requesting a new EHR, referenced by a name, in the audit log. |
committerId |
string |
no |
Store the information about the person requesting a new EHR, referenced by an ID, in the audit log. |
POST https://sandbox.better.care/ehr/rest/v1/?subjectId=123&subjectNamespace=emrPatientId
{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/aaa97f1b-59d1-4313-9c71-3d4438a436e6"
},
"action": "CREATE",
"ehrId": "aaa97f1b-59d1-4313-9c71-3d4438a436e6"
}Check the ehrId attribute, where the newly created EHR ID is returned.
Store it, preferably in the Demographics entry for Samantha.
Imagine you created an EHR and forgot to store the EHR ID part? Something went wrong. Your DB failed. Network hiccup happened or similar. Practically speaking, that EHR ID is lost to you.
No worries, you can create a new one for a new patient.
But what if there was content linked to the existing EHR and you lost the information about who this EHR belongs to?! How to find it?!
Here subjectId and subjectNamespace come to play.
Each time you create an EHR, pass this two values in such a way that you can uniquely identify each EHR ID by them!
In our case we decided that Samanthas' ID (123) in the EMR system would do just fine.
You might use the Demographics entry ID, even though that is too tightly coupled and does not exist outside our scenario.
Depending on the country, her Insurance ID, Social Security Number, NHS Number or Personal Identification Number would do great.
In subjectNamespace describe your choice.
| subjectNamespace | subjectId |
|---|---|
insuranceId |
KZZ3464236 |
nhsNumber |
1234567890 |
national.identifier |
134-4x2-tta3 |
Find an EHR ID and get its state
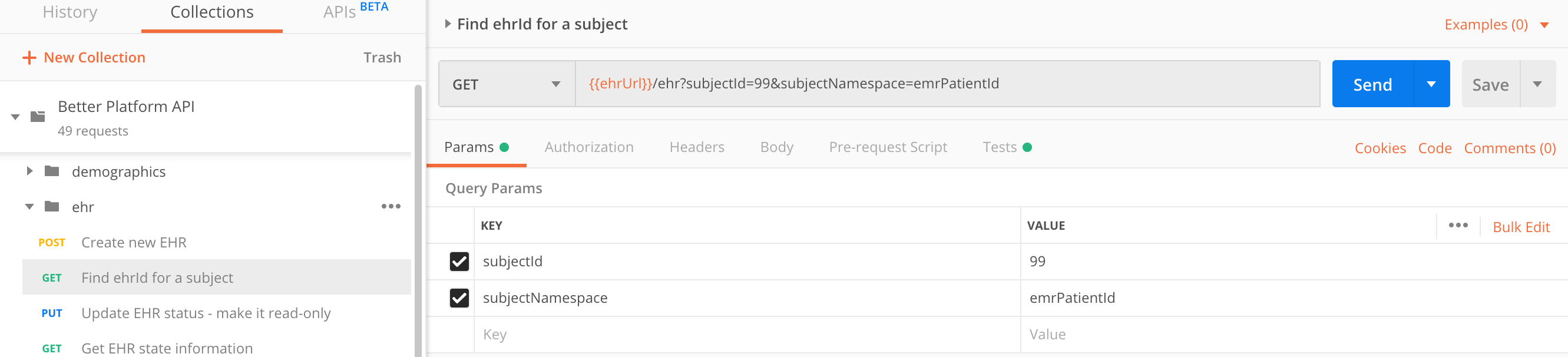
In case you lost the information which EHR ID belongs to which patient or if you decided not to manage this information in an external resource (such as Demographics), you can query for the EHR ID via the subjectId/subjectNamespace combination.
Next time Samantha comes for a check-up, your system needs to retrieve her EHR ID from the EHR Server.
| parameter | type | required | description |
|---|---|---|---|
subjectId |
string |
no |
A unique ID, that was matched with the patient/owner of the EHR. |
subjectNamespace |
string |
no |
Defines the domain of the |
GET https://sandbox.better.care/ehr/rest/v1/?subjectId=123&subjectNamespace=emrPatientId
{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/aaa97f1b-59d1-4313-9c71-3d4438a436e6"
},
"ehrStatus": {
"subjectId": "123",
"subjectNamespace": "emrPatientId",
"queryable": true,
"modifiable": true
},
"ehrId": "aaa97f1b-59d1-4313-9c71-3d4438a436e6"
}As you can notice, there are no clues about who this EHR belongs to, except for the external references subjectId and subjectNamespace.
| See subjectId/subjectNamespace section for more details. |
You get same response if you query the EHR Server for the status of a known EHR.
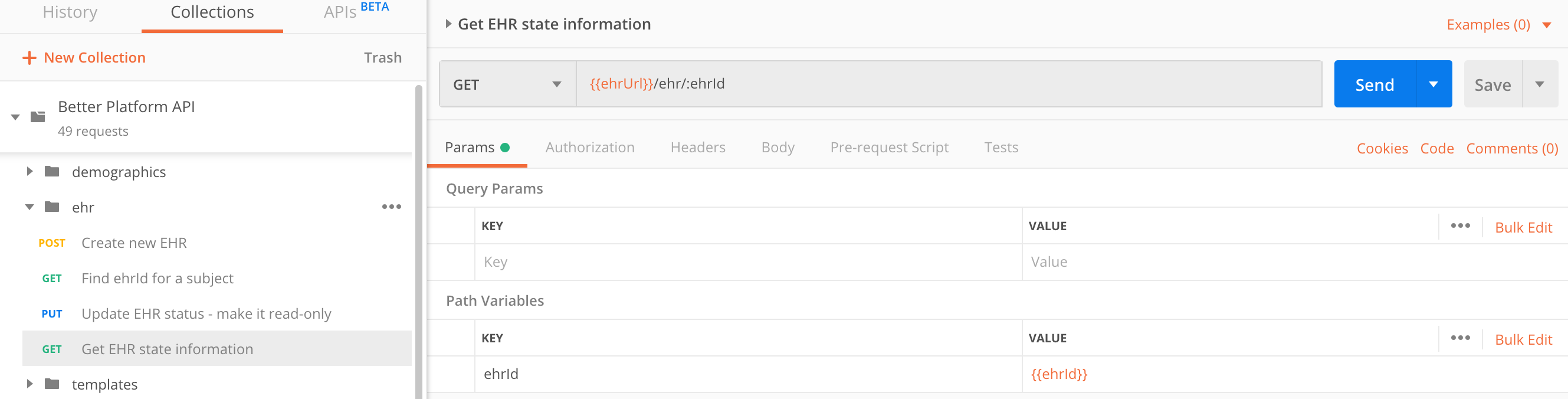
GET https://sandbox.better.care/ehr/rest/v1/aaa97f1b-59d1-4313-9c71-3d4438a436e6
ehrStatusAn EHR has two attributes, that describe its state:
| attribute | type | description |
|---|---|---|
queryable |
boolean |
When set to false, all queries explicitly referencing this specific EHR will fail. |
modifiable |
boolean |
When set to false, you cannot execute update/delete/create actions upon the EHR. |
|
You need to explicitly switch on the validation of the Non-queriable EHR data
Data stored in the compositions belonging to a non-queryable EHR will still be accessible through the AQL queries! |
If there are no results, the response with the HTTP status 204 will be returned.
Endpoint /template
In order to execute create or delete operations on a template you need special privileges.
Leave that to the administrators, try something different. Two basic operations you are likely to use the most are:
-
get a list of installed templates,
-
get a sample composition for an installed template.
Get a list of installed templates
Get a list of all installed templates and the date of the last update. Templates are not versioned on the EHR Server, only the current template for each of the names listed exists and is used for validation.
You can also search for specific templates according to their tag:values pairs or only tag values.
See Tag your templates section for more information on tags handling.
| parameter | type | required | description |
|---|---|---|---|
tags |
string |
no |
Pass a pipe ( |
search |
string |
no |
Pass a string to search for in the tag values.
Use |
GET https://sandbox.better.care/ehr/rest/v1/template
{
"templates": [
{
"templateId": "Allergies",
"createdOn": "2018-10-15T07:46:36.917Z"
},
{
"templateId": "Clinical Report",
"createdOn": "2018-10-15T07:46:36.954Z"
},
{
"templateId": "Encounter",
"createdOn": "2018-10-15T07:46:37.057Z"
},
{
"templateId": "Laboratory Report",
"createdOn": "2018-10-15T07:46:37.089Z"
},
{
"templateId": "Medical Diagnosis",
"createdOn": "2018-10-15T07:46:37.113Z"
},
{
"templateId": "Medications",
"createdOn": "2018-10-15T07:46:37.138Z"
},
{
"templateId": "Radiology Imaging Report",
"createdOn": "2018-10-15T07:46:37.175Z"
},
{
"templateId": "Vital Signs",
"createdOn": "2018-10-15T07:46:37.238Z"
}
]
}To list all templates with a tag platform and value mobile:
GET https://sandbox.better.care/ehr/rest/v1/template?tags=platform:mobile
To list all templates with a tag platform and tag ios:
GET https://sandbox.better.care/ehr/rest/v1/template?tags=platform:mobile&tags=ios
To list all templates with a tag platform and (tag ios OR android):
GET https://sandbox.better.care/ehr/rest/v1/template?tags=platform:mobile&tags=ios|android
To list all templates with mob substring in tag values:
GET https://sandbox.better.care/ehr/rest/v1/template?search=*mob*
Create a sample composition
No two templates are alike. The composition structure follows the template structure, which is set during the modelling process and you cannot change or influence it later.
Even if two different templates use the same blood pressure systolic field, it could be located in two different locations, thus, a different path would represent it.
To see how your model translates into an actual composition, use the template/<templateId>/example method to get a populated composition to learn from.
A generated composition is populated with dummy data, but it can be immediately sent to the EHR Server, since the dummy data falls within any existing restrictions.
| parameter | type | required | description |
|---|---|---|---|
format |
string |
no, |
Choose between:
|
exampleFilter |
string |
no, |
Choose between:
|
GET
https://sandbox.better.care/ehr/rest/v1/template/Vital%20Signs/example?format=FLAT&exampleFilter=INPUT
{
"ctx/language": "en",
"ctx/territory": "US",
"vital_signs/body_temperature:0/any_event:0/temperature|magnitude": 13.7,
"vital_signs/body_temperature:0/any_event:0/temperature|unit": "°C",
"vital_signs/body_temperature:0/any_event:0/body_exposure|code": "at0033",
"vital_signs/body_temperature:0/any_event:0/description_of_thermal_stress": "Description of thermal stress",
"vital_signs/body_temperature:0/site_of_measurement|code": "at0024",
"vital_signs/blood_pressure:0/any_event:0/systolic|magnitude": 242,
"vital_signs/blood_pressure:0/any_event:0/systolic|unit": "mm[Hg]",
"vital_signs/blood_pressure:0/any_event:0/diastolic|magnitude": 496,
"vital_signs/blood_pressure:0/any_event:0/diastolic|unit": "mm[Hg]",
"vital_signs/blood_pressure:0/cuff_size|code": "at1008",
"vital_signs/blood_pressure:0/location/location_of_measurement|code": "at1053",
"vital_signs/blood_pressure:0/device/device_name": "Device name",
"vital_signs/blood_pressure:0/device/description": "Description",
"vital_signs/blood_pressure:0/device/type": "Type",
"vital_signs/blood_pressure:0/device/manufacturer": "Manufacturer"
}Both carry same information, same level of details, it’s up to you to choose one.
In general FLAT is more usable when users manually edit it in text editor for example to prepare a template where placeholder values would be replaced by real values. Structured format on the other hand is more suitable when it’s manipulated programmatically - Better Platform’s form renderer for example uses structured format.
{
"ctx/language": "en",
"ctx/territory": "US",
"vital_signs": {
"body_temperature": [
{
"any_event": [
{
"temperature": [
{
"|magnitude": 13.7,
"|unit": "°C"
}
],
"body_exposure": [
{
"|code": "at0033"
}
],
"description_of_thermal_stress": [
"Description of thermal stress"
]
}
],
"site_of_measurement": [
{
"|code": "at0024"
}
]
}
],
"blood_pressure": [
{
"any_event": [
{
"systolic": [
{
"|magnitude": 242,
"|unit": "mm[Hg]"
}
],
"diastolic": [
{
"|magnitude": 496,
"|unit": "mm[Hg]"
}
]
}
],
"cuff_size": [
{
"|code": "at1008"
}
],
"location": [
{
"location_of_measurement": [
{
"|code": "at1053"
}
]
}
],
"device": [
{
"device_name": [
"Device name"
],
"description": [
"Description"
],
"type": [
"Type"
],
"manufacturer": [
"Manufacturer"
]
}
]
}
]
}
}Template fields mapping
In an openEHR model the fields are defined by an AQL path, but their actual location in the composition is defined in the template by a web template path.
Queries address fields by their AQL paths, compositions use the web template paths.
In order to get a mapping between the two, use the following request.
Include template ID in the request path:
GET https://sandbox.better.care/ehr/rest/v1/template/<templateId>/fields
Response is formed in an array of items with the format:
{
"name" : "<field name>",
"wtPath" : "<web template path>",
"aqlPath" : "<AQL path>"
}
GET https://sandbox.better.care/ehr/rest/v1/template/Vital%20Signs/fields
[
{
"name": "Start_time",
"wtPath": "vital_signs/context/start_time",
"aqlPath": "/context/start_time"
},
{
"name": "Setting",
"wtPath": "vital_signs/context/setting",
"aqlPath": "/context/setting"
},
{
"name": "Temperature",
"wtPath": "vital_signs/body_temperature/any_event/temperature",
"aqlPath": "/content[openEHR-EHR-OBSERVATION.body_temperature.v1]/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value"
},
{
"name": "Body exposure",
"wtPath": "vital_signs/body_temperature/any_event/body_exposure",
"aqlPath": "/content[openEHR-EHR-OBSERVATION.body_temperature.v1]/data[at0002]/events[at0003]/state[at0029]/items[at0030]/value"
},
{
"name": "Description of thermal stress",
"wtPath": "vital_signs/body_temperature/any_event/description_of_thermal_stress",
"aqlPath": "/content[openEHR-EHR-OBSERVATION.body_temperature.v1]/data[at0002]/events[at0003]/state[at0029]/items[at0041]/value"
},
:
:
:
{
"name": "vital_signs/category",
"wtPath": "vital_signs/category",
"aqlPath": "/category"
}
]Tag your templates
You can tag the templates for organizational purposes. Each tag can also have a value associated.
PUT https://sandbox.better.care/ehr/rest/v1/template/Vital%20Signs/tag?tags=ward:icu
HTTP code 204 marks a successful request.
Add multiple values for same tag:
PUT https://sandbox.better.care/ehr/rest/v1/template/Vital%20Signs/tag?tags=ward:icu|ward:emergency
Add multiple tags:
PUT https://sandbox.better.care/ehr/rest/v1/template/Vital%20Signs/tag?tags=ward:icu|importance:high
Add a tag without a value:
PUT https://sandbox.better.care/ehr/rest/v1/template/Vital%20Signs/tag?tags=wound
Endpoint /composition
Once you have an EHR open, you know its EHR ID and a valid template is present on the EHR Server, you can start storing data compositions in that EHR and retrieving them from it.
Each time you try storing a composition in the EHR Server, it is validated against a template you choose, for structure and values.
By now you know how to get a sample composition for your template, where you start with a skeleton structure to fill in with data.
Composition ID
It uniquely identifies an EHR Server document.
It has three parts, delimited with a double column (::):
-
versioned composition identifier (does not change between versions),
-
system id,
-
version.
When a document is referenced only by the first part (versioned composition identifier), it is assumed to be the latest version of it.
Create a composition
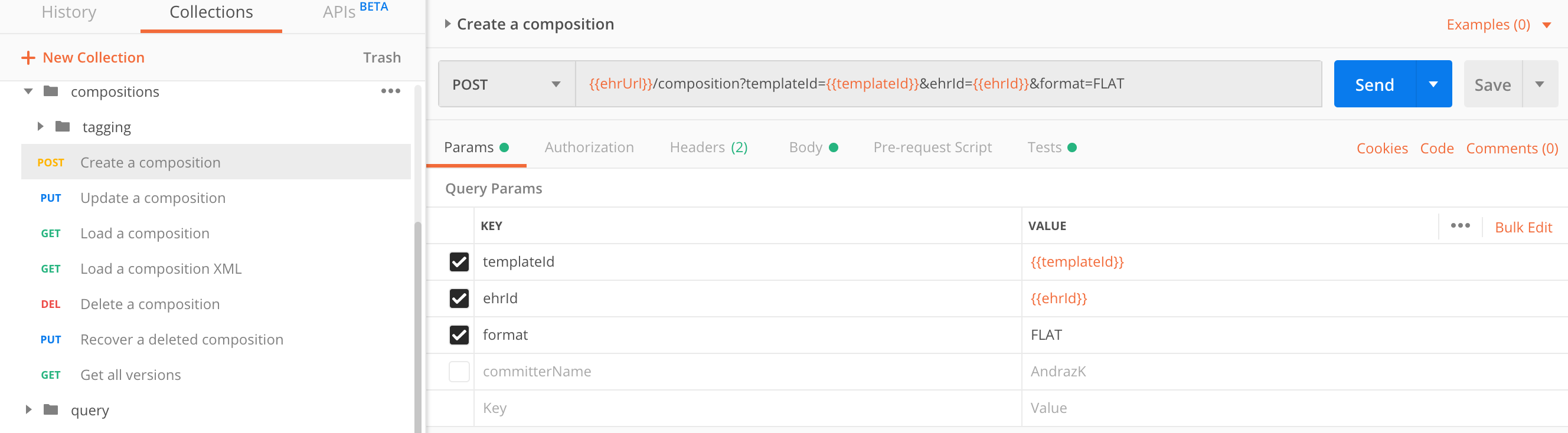
Put the composition in requests body.
| parameter | type | required | description |
|---|---|---|---|
templateId |
string |
yes |
Unless the |
ehrId |
string |
yes |
The composition must be linked to a specific EHR, identified by this EHR ID. |
format |
string |
yes |
Request body contents must be in |
committerName |
string |
no |
If passed, the value will override any data set in the composition |
comment |
string |
no |
This value is stored as a comment in the audit log and is not part of the composition itself. |
|
Set the request headers to |
Let’s try something simple.
POST https://sandbox.better.care/ehr/rest/v1/composition?templateId=Vital%20Signs&ehrId=aaa97f1b-59d1-4313-9c71-3d4438a436e6&format=FLAT
{
"ctx/language": "en",
"ctx/territory": "US",
"vital_signs/body_temperature:0/any_event:0/temperature|magnitude": 73.2,
"vital_signs/body_temperature:0/any_event:0/temperature|unit": "°C",
"vital_signs/body_temperature:0/site_of_measurement|code": "at0027"
}{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1"
},
"action": "CREATE",
"compositionUid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1"
}You just created your first valid composition!
And as you can see you got its composition ID in the compositionUid attribute.
Let’s check the stored composition now!
Load a composition
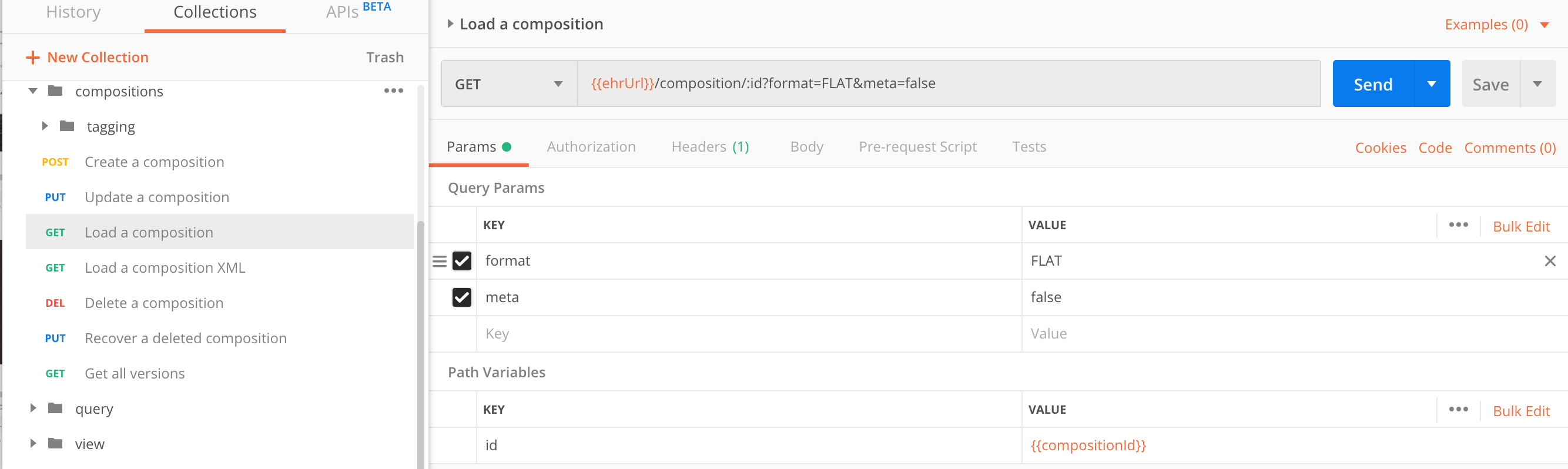
A composition is not stored as a text document, therefore we must serialize it in one of the formats for presentation.
If besides the clinical data you also need the composition meta-data, you can get that as a composition wrapper.
| parameter | type | required | description |
|---|---|---|---|
format |
string |
no, |
Composition can be serialized to |
meta |
boolean |
no, |
Wrap the composition in meta-data, such as EHR ID, composition ID, template ID it was checked with, status etc. |
Pass the compositionUid as a path parameter:
GET https://sandbox.better.care/ehr/rest/v1/composition/<compositionUid>
GET https://sandbox.better.care/ehr/rest/v1/composition/00bf6f8a-62d7-423a-8c3e-3156cef87824::andrazk-demo.sandbox.com::1?meta=true&format=FLAT
{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1"
},
"compositionUid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1",
"format": "FLAT",
"templateId": "Vital Signs",
"composition": {
"vital_signs/_uid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1",
"vital_signs/language|code": "en",
"vital_signs/language|terminology": "ISO_639-1",
"vital_signs/territory|code": "US",
"vital_signs/territory|terminology": "ISO_3166-1",
"vital_signs/context/start_time": "2019-05-06T09:27:31.305+02:00",
"vital_signs/context/setting|code": "238",
"vital_signs/context/setting|value": "other care",
"vital_signs/context/setting|terminology": "openehr",
"vital_signs/body_temperature:0/any_event:0/temperature|magnitude": 73.2,
"vital_signs/body_temperature:0/any_event:0/temperature|unit": "°C",
"vital_signs/body_temperature:0/any_event:0/time": "2019-05-06T09:27:31.305+02:00",
"vital_signs/body_temperature:0/site_of_measurement|code": "at0027",
"vital_signs/body_temperature:0/site_of_measurement|value": "Urinary bladder",
"vital_signs/body_temperature:0/site_of_measurement|terminology": "local",
"vital_signs/body_temperature:0/language|code": "en",
"vital_signs/body_temperature:0/language|terminology": "ISO_639-1",
"vital_signs/body_temperature:0/encoding|code": "UTF-8",
"vital_signs/body_temperature:0/encoding|terminology": "IANA_character-sets",
"vital_signs/category|code": "433",
"vital_signs/category|value": "event",
"vital_signs/category|terminology": "openehr",
"vital_signs/composer|name": "andrazk"
},
"deleted": false,
"lastVersion": true,
"ehrId": "aaa97f1b-59d1-4313-9c71-3d4438a436e6",
"lifecycleState": "COMPLETE"
}| attribute | type | description |
|---|---|---|
meta |
object |
Can contain:
|
compositionUid |
string |
Unique full composition identifier. |
templateId |
string |
Name of the template that was used to store the composition. |
deleted |
boolean |
If set to |
lastVersion |
boolean |
Set to |
ehrId |
string |
Represents the EHR to which this composition belongs. |
{
"vital_signs/_uid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1",
"vital_signs/language|code": "en",
"vital_signs/language|terminology": "ISO_639-1",
"vital_signs/territory|code": "US",
"vital_signs/territory|terminology": "ISO_3166-1",
"vital_signs/context/start_time": "2019-05-06T09:27:31.305+02:00",
"vital_signs/context/setting|code": "238",
"vital_signs/context/setting|value": "other care",
"vital_signs/context/setting|terminology": "openehr",
"vital_signs/body_temperature:0/any_event:0/temperature|magnitude": 73.2,
"vital_signs/body_temperature:0/any_event:0/temperature|unit": "°C",
"vital_signs/body_temperature:0/any_event:0/time": "2019-05-06T09:27:31.305+02:00",
"vital_signs/body_temperature:0/site_of_measurement|code": "at0027",
"vital_signs/body_temperature:0/site_of_measurement|value": "Urinary bladder",
"vital_signs/body_temperature:0/site_of_measurement|terminology": "local",
"vital_signs/body_temperature:0/language|code": "en",
"vital_signs/body_temperature:0/language|terminology": "ISO_639-1",
"vital_signs/body_temperature:0/encoding|code": "UTF-8",
"vital_signs/body_temperature:0/encoding|terminology": "IANA_character-sets",
"vital_signs/category|code": "433",
"vital_signs/category|value": "event",
"vital_signs/category|terminology": "openehr",
"vital_signs/composer|name": "andrazk"
}In vs. out
Now, compare what we sent into the repository and what we got from it.
There seems to be more data available. What happened?
-
Coded data fields are paired and looked-up into terminologies, now it contains not just the code but also a human readable value:
"vital_signs/body_temperature:0/site_of_measurement|code": "at0027", "vital_signs/body_temperature:0/site_of_measurement|value": "Urinary bladder", "vital_signs/body_temperature:0/site_of_measurement|terminology": "local",
-
Some fields have attributes originating in the Reference Model, like the Event
time. If there is no explicit value passed, it assumes the value of the current EHR Server time:
"vital_signs/body_temperature:0/any_event:0/time": "2019-05-06T09:27:31.305+02:00",
-
Some input values are translated to the output, according to the Web Template specifications. An example of this would be
ctx/composer_nameon the input, which is translated to"vital_signs/composer|name": "andrazk"on the output.
Update a composition
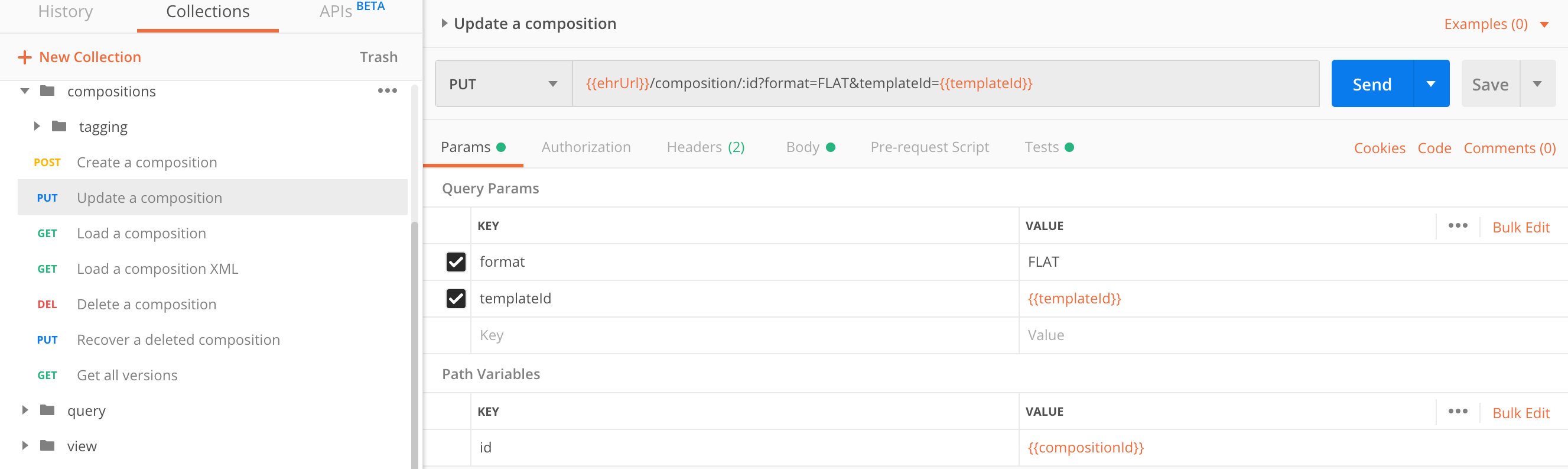
A composition is an immutable object which you cannot edit, but you can create a new version of it. When there are multiple composition versions, only the last one is active and its data can be captured with queries.
You can update only the last - currently active composition version.
Any of the compositions can be retrieved from the EHR Server, event the previous versions, when explicitly requesting them via path parameter compositionUid parameter.
We made a mistake saving the patient body temperature and entered the value of 73.2 degrees instead of 37.2. Let’s correct that.
| parameter | type | required | description |
|---|---|---|---|
format |
string |
yes |
Composition can be serialized to |
templateId |
string |
yes |
Unless the |
Request body contains a composition, same as when you create a new composition, containing the updates. In this one we corrected the 73.2 temperature value to 37.2:
{
"ctx/language": "en",
"ctx/territory": "US",
"vital_signs/body_temperature:0/any_event:0/temperature|magnitude": 37.2,
"vital_signs/body_temperature:0/any_event:0/temperature|unit": "°C",
"vital_signs/body_temperature:0/site_of_measurement|code": "at0027"
}Pass the compositionUid as a path parameter:
PUT https://sandbox.better.care/ehr/rest/v1/composition/<compositionUid>
PUT https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1?templateId=Vital%20Signs&format=FLAT
{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::2"
},
"action": "UPDATE",
"compositionUid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::2"
}You can see the version updated to 2.
What else?
When you request an updated composition together with its meta-data, you can see the previous version referenced in meta.precedingHref attribute:
{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::2",
"precedingHref": "https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1"
},
"compositionUid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::2",
"format": "FLAT",
"templateId": "Vital Signs",
"composition": {
"vital_signs/_uid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::2",
"vital_signs/language|code": "en",
"vital_signs/language|terminology": "ISO_639-1",
"vital_signs/territory|code": "US",
"vital_signs/territory|terminology": "ISO_3166-1",
"vital_signs/context/start_time": "2019-05-06T11:15:07.421+02:00",
"vital_signs/context/setting|code": "238",
"vital_signs/context/setting|value": "other care",
"vital_signs/context/setting|terminology": "openehr",
"vital_signs/body_temperature:0/any_event:0/temperature|magnitude": 37.2,
"vital_signs/body_temperature:0/any_event:0/temperature|unit": "°C",
"vital_signs/body_temperature:0/any_event:0/time": "2019-05-06T11:15:07.421+02:00",
"vital_signs/body_temperature:0/site_of_measurement|code": "at0027",
"vital_signs/body_temperature:0/site_of_measurement|value": "Urinary bladder",
"vital_signs/body_temperature:0/site_of_measurement|terminology": "local",
"vital_signs/body_temperature:0/language|code": "en",
"vital_signs/body_temperature:0/language|terminology": "ISO_639-1",
"vital_signs/body_temperature:0/encoding|code": "UTF-8",
"vital_signs/body_temperature:0/encoding|terminology": "IANA_character-sets",
"vital_signs/category|code": "433",
"vital_signs/category|value": "event",
"vital_signs/category|terminology": "openehr",
"vital_signs/composer|name": "andrazk"
},
"deleted": false,
"lastVersion": true,
"ehrId": "aaa97f1b-59d1-4313-9c71-3d4438a436e6",
"lifecycleState": "COMPLETE"
}GET https://sandbox.better.care/rest/v1/composition/00bf6f8a-62d7-423a-8c3e-3156cef87824::andrazk-demo.sandbox.com::1?meta=true&format=FLAT
{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1",
"nextHref": "https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::2"
},
"compositionUid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1",
"format": "FLAT",
"templateId": "Vital Signs",
"composition": {
"vital_signs/_uid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::1",
"vital_signs/language|code": "en",
"vital_signs/language|terminology": "ISO_639-1",
"vital_signs/territory|code": "US",
"vital_signs/territory|terminology": "ISO_3166-1",
"vital_signs/context/start_time": "2019-05-06T09:27:31.305+02:00",
"vital_signs/context/setting|code": "238",
"vital_signs/context/setting|value": "other care",
"vital_signs/context/setting|terminology": "openehr",
"vital_signs/body_temperature:0/any_event:0/temperature|magnitude": 73.2,
"vital_signs/body_temperature:0/any_event:0/temperature|unit": "°C",
"vital_signs/body_temperature:0/any_event:0/time": "2019-05-06T09:27:31.305+02:00",
"vital_signs/body_temperature:0/site_of_measurement|code": "at0027",
"vital_signs/body_temperature:0/site_of_measurement|value": "Urinary bladder",
"vital_signs/body_temperature:0/site_of_measurement|terminology": "local",
"vital_signs/body_temperature:0/language|code": "en",
"vital_signs/body_temperature:0/language|terminology": "ISO_639-1",
"vital_signs/body_temperature:0/encoding|code": "UTF-8",
"vital_signs/body_temperature:0/encoding|terminology": "IANA_character-sets",
"vital_signs/category|code": "433",
"vital_signs/category|value": "event",
"vital_signs/category|terminology": "openehr",
"vital_signs/composer|name": "andrazk"
},
"deleted": false,
"lastVersion": false,
"ehrId": "aaa97f1b-59d1-4313-9c71-3d4438a436e6",
"lifecycleState": "COMPLETE"
}Observe the following attributes:
-
meta.nextHrefexists, because this composition was updated, -
lastVersionis false, because this is not the last of the composition versions.
Q and A
-
When do you want to update a composition?
When it contains an error in the data.
-
What if I stored a measurement in a single document and now I got more linked measurements?
Create new compositions, link them with references. There is not much difference reading data from a single or multiple documents.
-
Will an update break internal references?
No, since you can reference using a composition identifier, which does not change when updating a composition. Only the version is changed.
-
How do I get previous versions of a composition?
Request the composition with a version specified. It still exists in the EHR Server, but its state is
archived. -
Will an update affect the queries?
The data from the archived compositions is not included in the AQL queries result, unless you explicitly specify that criteria in the query.
Delete a composition
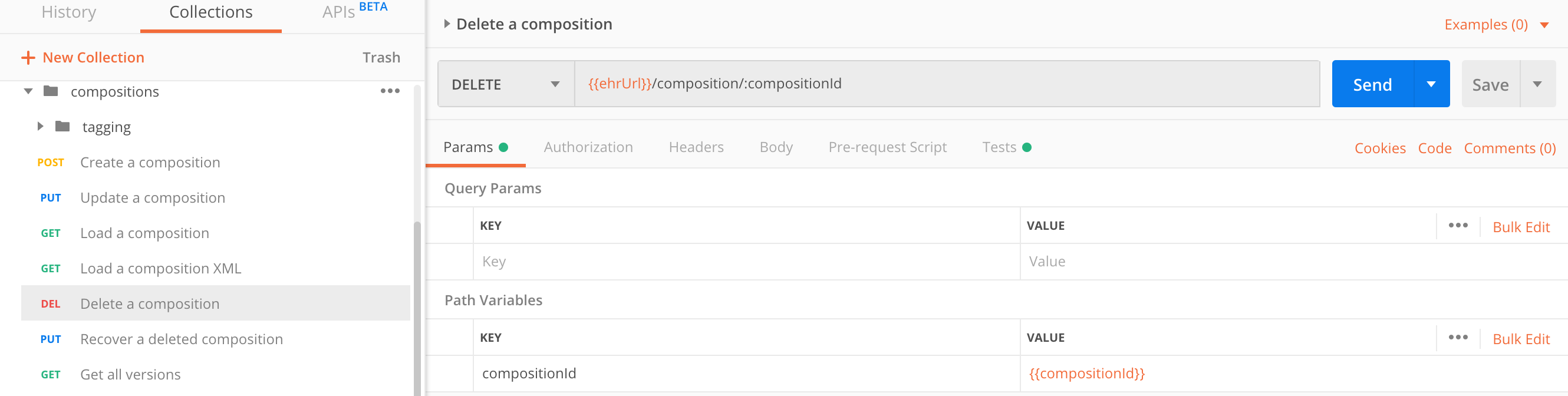
A deleted composition is not physically removed from the repository but is only marked as deleted.
All its contents are not included in the AQL queries result, unless you explicitly specify that criteria in the query.
You can delete only the last - currently active composition version. Deleting a compositions is understood as updating its state.
DELETE https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::2
{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::3"
},
"action": "DELETE",
"compositionUid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::3"
}The composition version 3 is therefore empty - deleted, which you can check by accessing its meta-data.
Restore a deleted composition
You can only restore a deleted composition if it is also the last version.
The restore action will create a new composition, version 4.
PUT https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::3/restore
{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/composition/a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::4"
},
"action": "EXECUTE",
"compositionUid": "a0ecfaea-097e-48bb-9f79-8bde51fa2634::andrazk-demo.sandbox.com::4"
}Endpoint /query
To access specific data, not necessarily the complete compositions, you can send AQL queries towards the EHR Server, which then serves the serialized result set.
We will discuss two types of requests:
-
simple query request to execute a query via a HTTP GET request, which you can test from a browser,
-
advanced query request, which is a parameter driven HTTP POST request.
Simple query request
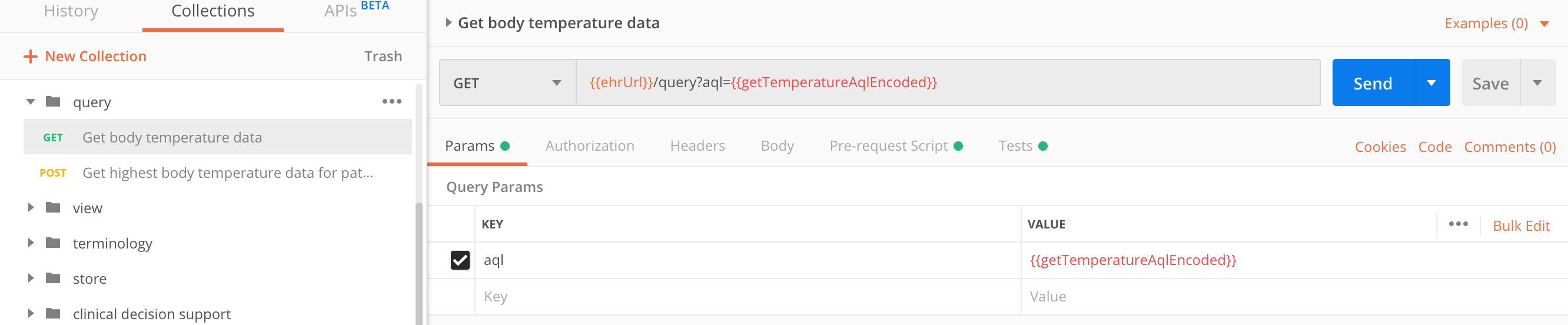
| parameter | type | required | description |
|---|---|---|---|
aql |
string |
yes |
URL-encoded AQL query statement. |
The query we want to execute is querying the body temperature data:
select o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as bt
from EHR e contains COMPOSITION c
contains OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v1]
offset 0 limit 100GET https://sandbox.better.care/ehr/rest/v1/query/?aql=select%20c/uid/value%20as%20cid,%20o/data%5Bat0002%5D/events%5Bat0003%5D/data%5Bat0001%5D/items%5Bat0004%5D/value%20as%20bt%20from%20EHR%20e%20contains%20COMPOSITION%20c%20contains%20OBSERVATION%20o%5BopenEHR-EHR-OBSERVATION.body_temperature.v1%5D%20%20limit%203
{{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/query/?aql=select%20c/uid/value%20as%20cid,%20o/data%5Bat0002%5D/events%5Bat0003%5D/data%5Bat0001%5D/items%5Bat0004%5D/value%20as%20bt%20from%20EHR%20e%20contains%20COMPOSITION%20c%20contains%20OBSERVATION%20o%5BopenEHR-EHR-OBSERVATION.body_temperature.v1%5D%20%20limit%203"
},
"aql": "select c/uid/value as cid, o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as bt from EHR e contains COMPOSITION c contains OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v1] limit 3",
"executedAql": "select c/uid/value as cid, o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as bt from EHR e contains COMPOSITION c contains OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v1] limit 3",
"resultSet": [
{
"cid": "7477ae7d-f295-4fbf-89ba-6b94ebb2aa72::andrazk-demo.sandbox.com::1",
"bt": {
"@class": "DV_QUANTITY",
"magnitude": 36.8,
"units": "°C",
"precision": 1
}
},
{
"cid": "a500a355-dba0-4b01-86ca-ec19acc25c7f::andrazk-demo.sandbox.com::1",
"bt": {
"@class": "DV_QUANTITY",
"magnitude": 37.4,
"units": "°C",
"precision": 1
}
},
{
"cid": "e59e5a39-9d8d-44f9-b905-a30f43a2c0e0::andrazk-demo.sandbox.com::1",
"bt": {
"@class": "DV_QUANTITY",
"magnitude": 37.1,
"units": "°C",
"precision": 1
}
}
]
}The data you queried for is in the resultSet attribute.
Advanced query requests
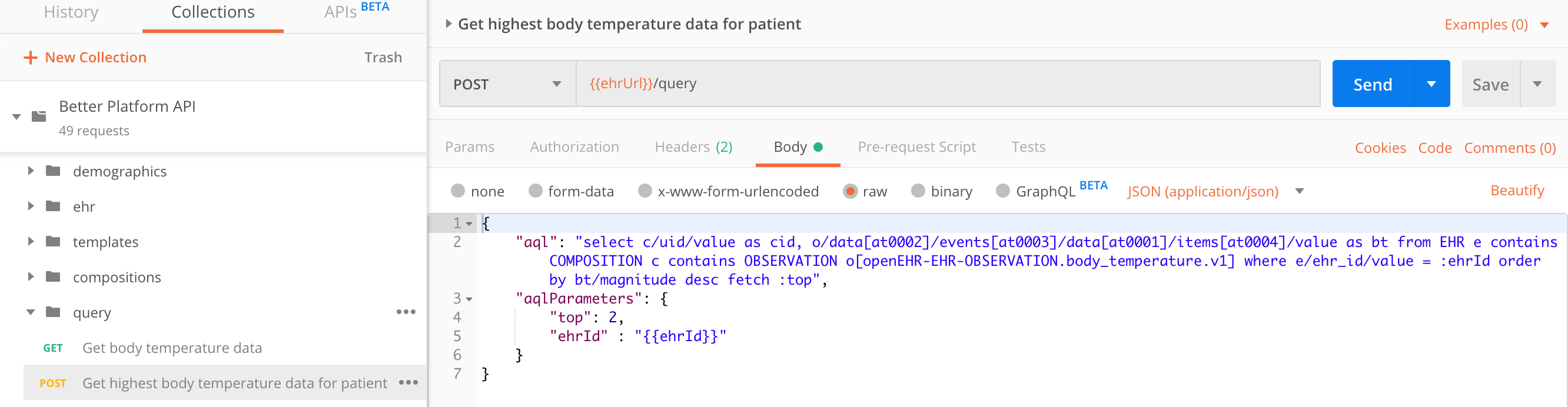
The advantage of the advanced query over the simple one is that the query length itself is not limited by the maximum URL length (sometimes as little as 1k characters) and that the parameters are separated from the query itself, which makes query management easier.
In this example we are querying the top 2 highest body temperature entries for a specific patient.
select c/uid/value as cid,
o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as bt
from EHR e
contains COMPOSITION c
contains OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v1]
where e/ehr_id/value = :ehrId
order by bt/magnitude desc
fetch :topPOST https://sandbox.better.care/ehr/rest/v1/query
{
"aql": "select c/uid/value as cid, o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as bt from EHR e contains COMPOSITION c contains OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v1] where e/ehr_id/value = :ehrId order by bt/magnitude desc fetch :top",
"aqlParameters": {
"top": 2,
"ehrId" : "aaa97f1b-59d1-4313-9c71-3d4438a436e6"
}
}{
"meta": {
"href": "https://sandbox.better.care/ehr/rest/v1/query/"
},
"aql": "select c/uid/value as cid, o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as bt from EHR e contains COMPOSITION c contains OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v1] where e/ehr_id/value = :ehrId order by bt/magnitude desc fetch :top",
"executedAql": "select c/uid/value as cid, o/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value as bt from EHR e contains COMPOSITION c contains OBSERVATION o[openEHR-EHR-OBSERVATION.body_temperature.v1] where e/ehr_id/value = 'aaa97f1b-59d1-4313-9c71-3d4438a436e6' order by bt/magnitude desc fetch 2",
"aqlParameters": {
"top": 2,
"ehrId": "aaa97f1b-59d1-4313-9c71-3d4438a436e6"
},
"resultSet": [
{
"cid": "3ac587a5-cd60-45b8-8a21-92c6c398672a::andrazk-demo.sandbox.com::1",
"bt": {
"@class": "DV_QUANTITY",
"magnitude": 37.7,
"units": "°C",
"precision": 1
}
},
{
"cid": "00bf6f8a-62d7-423a-8c3e-3156cef87824::andrazk-demo.sandbox.com::1",
"bt": {
"@class": "DV_QUANTITY",
"magnitude": 37.2,
"units": "°C",
"precision": 1
}
}
]
}The data you queried for is in the resultSet attribute.
Endpoint /view
Having queries reside on client side is not always the best of options. You need to consider the software updates, with new logic, new data and it is on you to make it all work. Synchronized update to the clients and the server, supporting multiple versions etc.
Also, an AQL query request looks very much alike to a SQL query and sending those across the wire looks suspicious. Trigger happy firewalls and security devices might block such request under the suspicion of an SQL injection attack!
Hence, server based queries are the way to go, same as standard database views solve similar dilemmas.
Setup a view
Views are setup through EHR Server administration dashboard.
| Please see Better Platform: Developers guide, section _Views for detailed instructions on how to create a view. |
Access data through a view
Even though it is called a view, which we usually percept as a "static" representation of data, EHR Server view accepts parameters, therefore it behaves more as a stored procedure or prepared statement.
View parameters need to be declared in the view definition, but there are a couple parameters that are predefined and always passed to the view.
| parameters | type | description |
|---|---|---|
ehrId |
string |
Unique identifier, which is extracted from the request path, if it exists. If passed as a HTTP query parameter, it needs to be explicitly defined in the view parameters definition. |
offset |
integer, |
Defines the offset from the start of the result set, used for paging purposes. |
limit |
integer |
Limit the size of the result set. |
body_temperature definitionselect
a_a/data[at0002]/events[at0003]/time/value as time,
a_a/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value/magnitude as temperature,
a_a/data[at0002]/events[at0003]/data[at0001]/items[at0004]/value/units as unit
from EHR e
contains OBSERVATION a_a[openEHR-EHR-OBSERVATION.body_temperature.v1]
where e/ehr_id/value = :ehrId
<#if ctx.vars.from??>and a_a/data[at0002]/events[at0003]/time >= :from</#if>
<#if ctx.vars.to??>and a_a/data[at0002]/events[at0003]/time <= :to</#if>
order by time desc
offset :offset fetch :limitThis view is of type Freemarker AQL → JSON accepts two extra parameters:
| parameter | type | description |
|---|---|---|
from |
ISO8601 date-time string |
Set a result set start point in time. |
to |
ISO8601 date-time string |
Set a result set end point in time. |
Simple way of accessing view data
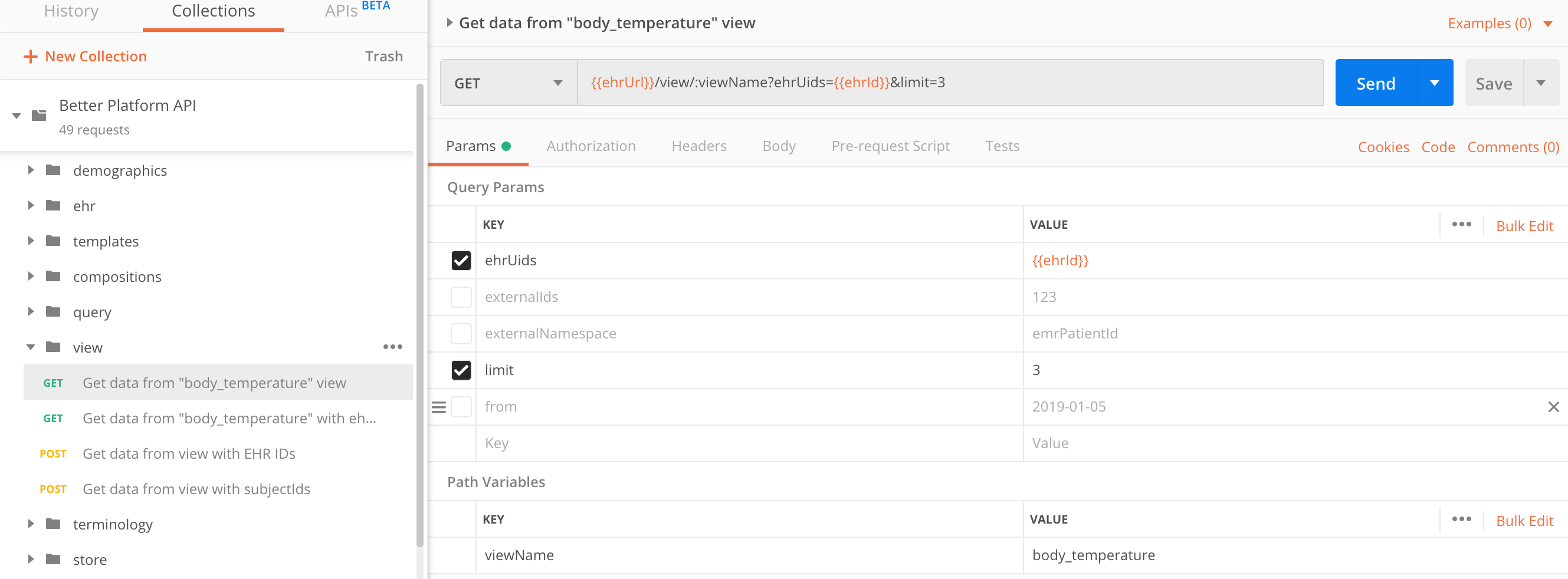
Call the view with its name:
GET https://sandbox.better.care/ehr/rest/v1/view/<viewName>
| parameter | type | required | description |
|---|---|---|---|
ehrUids |
string |
yes, if |
If you use the parameter |
subjectIds |
string |
yes, if |
A comma separated list of |
subjectNamespace |
string |
yes, if |
Define the domain of |
externalIds and externalNamespace are deprecated, but still accepted until next major release.
Please use subjectIds and subjectNamespace instead.
|
ehrUidsGET https://sandbox.better.care/ehr/rest/v1/view/body_temperature?ehrUids=aaa97f1b-59d1-4313-9c71-3d4438a436e6&limit=3
Alternative request, using subjectIds would be:
GET https://sandbox.better.care/ehr/rest/v1/view/body_temperature?subjectIds=123&subjectNamespace=emrPatientId&limit=3
See EHR creation for more details about subjectIds.
|
{
"aaa97f1b-59d1-4313-9c71-3d4438a436e6": [
{
"unit": "°C",
"temperature": 36.5,
"time": "2019-05-09T13:53:48.486+02:00"
},
{
"unit": "°C",
"temperature": 37.7,
"time": "2019-05-09T13:53:36.99+02:00"
},
{
"unit": "°C",
"temperature": 37.2,
"time": "2019-04-29T15:34:30.191+02:00"
}
]
}Response always groups the result sets by the EHR IDs:
{
"ehrId1" : [
{ ... },
{ ... }
],
"ehrId2" : [
{ ... },
{ ... }
]
}Get data for multiple comma separated EHR IDs:
GET https://sandbox.better.care/ehr/rest/v1/view/body_temperature?ehrUids=aaa97f1b-59d1-4313-9c71-3d4438a436e6,02e1aac1-ae7e-41e6-be9d-328855a51eeb&limit=3
{
"aaa97f1b-59d1-4313-9c71-3d4438a436e6": [
{
"unit": "°C",
"temperature": 36.5,
"time": "2019-05-09T13:53:48.486+02:00"
},
{
"unit": "°C",
"temperature": 37.7,
"time": "2019-05-09T13:53:36.99+02:00"
},
{
"unit": "°C",
"temperature": 37.2,
"time": "2019-04-29T15:34:30.191+02:00"
}
],
"02e1aac1-ae7e-41e6-be9d-328855a51eeb": [
{
"unit": "°C",
"temperature": 36.4,
"time": "2019-04-09T11:28:13.391+02:00"
}
]
}Value of limit applies for each of the EHR IDs separately.
Include ehrId in request path
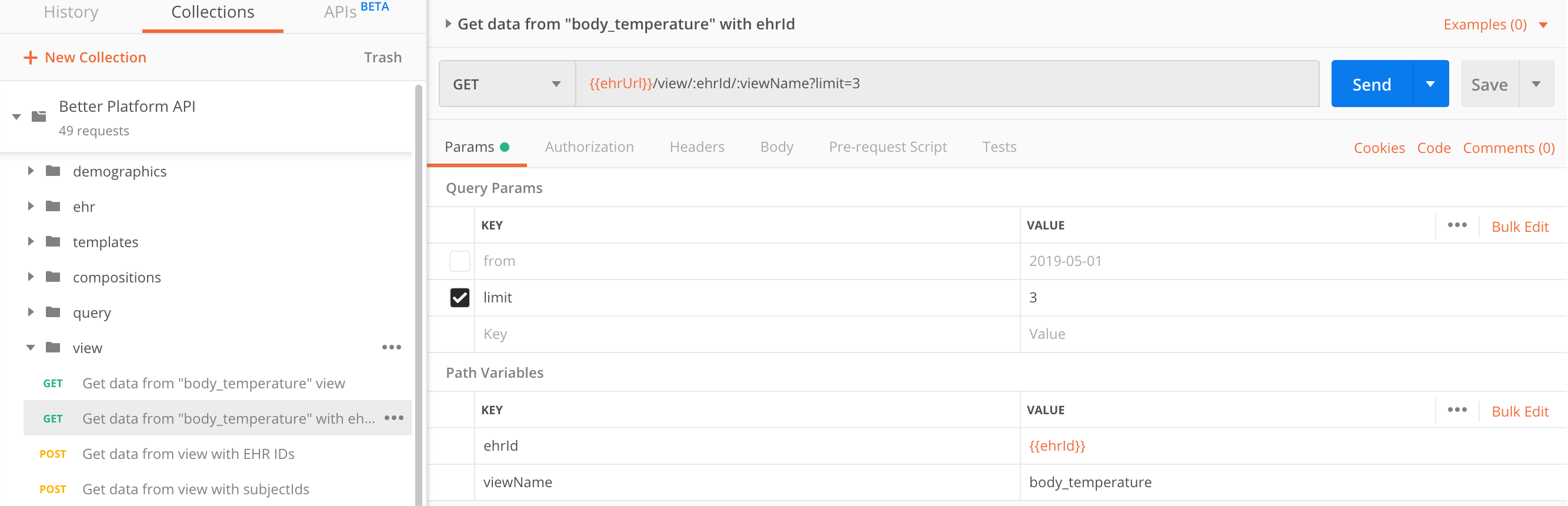
ehrUidsYou can invoke the same view call with the addition of a single EHR ID, incorporating it in the request path.
GET https://sandbox.better.care/ehr/rest/v1/view/<ehrUid>/<viewName>
ehrIdGET https://sandbox.better.care/ehr/rest/v1/view/aaa97f1b-59d1-4313-9c71-3d4438a436e6/body_temperature?limit=3
Advanced view call
Pass the view parameters through a HTTP POST call.
Only parameter is viewName in the request path.
POST https://sandbox.better.care/ehr/rest/v1/view/<viewName>
The list of EHR IDs is passed in the request body in format:
{
"ehrUids": [ "aaa97f1b-59d1-4313-9c71-3d4438a436e6", "02e1aac1-ae7e-41e6-be9d-328855a51eeb" ]
}Alternative method is to use subjectIds:
{
"subjectNamespace": "emrPatientId",
"subjectIds" : ["123"]
}
externalIds and externalNamespace are deprecated, but still accepted until next major release.
Please use subjectIds and subjectNamespace instead.
|
POST https://sandbox.better.care/ehr/rest/v1/view/body_temperature?limit=3
Same response as here.
Terminology API
Often you need access to specific terminology, its subset or you create your own and want to share it. Terminology server supports creating new terminologies and accessing the data, by directly uploading a terminology CSV file to the server and expose its data via a REST API.
You need special privileges to access the Terminology Server administrative API endpoint, where you can create, delete and update the terminologies.
Terminologies can be flat, where all entities are on the same level or hierarchical, where there are established relations parent/child between the terminology entities.
Administration
Administration endpoint has an admin prefix in the request path:
GET https://sandbox.better.care/terminology-adapter/rest/admin/terminology/codesystem/<codeSystem>
| See Terminology Server guide for all the details! |
Access the terminologies
All terminology related operations run on
https://sandbox.better.care/terminology-adapter/rest/terminology/ endpoint.
Requests support paging and localization with the following parameters:
| parameter | type | description |
|---|---|---|
page |
integer |
Page of results to return.
First page is number |
pagesize |
integer |
Maximum number of entities in a page. |
language |
string |
Send the ISO639-1 language code for response localization. |
Get a terminologies list
First of all, let’s get a list of all installed terminologies.
| parameter | type | required | description |
|---|---|---|---|
matchvalue |
string |
no |
Filter results by this value, contained in terminology |
match |
string |
no, unless |
It sets the way of performing a match, which is case insensitive. Can be one of the following values:
|
GET https://sandbox.better.care/terminology-adapter/rest/terminology/codesystems
{
"items": [
{
"code": "Pk-Nanda",
"description": "Pk-Nanda",
"version": "unversioned",
"languages": [
""
],
"lastModified": "2016-09-08T07:50:16Z"
},
{
"code": "Medications",
"description": "Medications",
"version": "unversioned",
"languages": [
""
],
"lastModified": "2016-09-08T07:50:16Z"
},
{
"code": "ICD10",
"description": "ICD10",
"version": "unversioned",
"languages": [
""
],
"lastModified": "2016-09-08T07:50:16Z"
},
{
"code": "LAB-PK",
"description": "LAB-PK",
"version": "unversioned",
"languages": [
""
],
"lastModified": "2016-09-08T07:50:16Z"
}
],
"lastPage": true
}Properties explained:
-
[item].version- versions are not supported yet, therefore this value is alwaysunversioned -
[item].languages- a list of languages the terminology has entries for. If none are defined, localization is not supported and the default values will be used. -
lastPage- flag specifying this is the results last page
Retrieve terminology data
To retrieve terminology data you usually query the data with a set of criteria in terms of content, relation or actual size of the result set.
| parameter | type | required | description |
|---|---|---|---|
entity |
string |
no |
Code(s) of the entity to be returned. Multiple values can be comma delimited or specified as multiple 'entity' parameters. |
filtercomponent |
string |
no |
Attributes under which to search.
Can have more than one value.
Valid values are: |
validAt |
ISO8601 date-time string |
no |
Returns codes that are valid at this instant of time. Default value equals to current server time. |
matchvalue |
string |
no |
Filter results by this value, contained in terminology |
match |
string |
no, unless |
It sets the way of performing a match, which is case insensitive. Can be one of the following values:
|
Do not forget about the common terminology parameters.
Pass the codeSystem as a path parameter:
GET https://sandbox.better.care/terminology-adapter/rest/terminology/codesystem/<codeSystem>/entities
Our testing code system will be ICD10.
GET https://sandbox.better.care/terminology-adapter/rest/terminology/codesystem/ICD10/entities?pagesize=5
{
"items": [
{
"code": "A00",
"description": "Cholera",
"childrenCount": 3
},
{
"code": "A000",
"description": "Cholera due to Vibrio cholerae 01, biovar cholerae",
"parent": "A00"
},
{
"code": "A001",
"description": "Cholera due to Vibrio cholerae 01, biovar eltor",
"parent": "A00"
},
{
"code": "A009",
"description": "Cholera, unspecified",
"parent": "A00"
},
{
"code": "A01",
"description": "Typhoid and paratyphoid fevers",
"childrenCount": 5
}
],
"lastPage": false
}Properties explained:
-
childrenCount- present only if the entity has any child entities -
parent- present only if this is a child entity, does not exist on root level entites.
Get an entity from ICD10 terminology with a code A00:
GET https://sandbox.better.care/terminology-adapter/rest/terminology/codesystem/ICD10/entity/A00`
{
"code": "A00",
"description": "Cholera",
"childrenCount": 3
}Get an entity and its children from ICD10 terminology, in this case an entity with the code A00:
GET https://sandbox.better.care/terminology-adapter/rest/terminology/codesystem/ICD10/entities?filtercomponent=parent&matchvalue=A00
{
"items": [
{
"code": "A00",
"description": "Cholera",
"childrenCount": 3
},
{
"code": "A000",
"description": "Cholera due to Vibrio cholerae 01, biovar cholerae",
"parent": "A00"
},
{
"code": "A001",
"description": "Cholera due to Vibrio cholerae 01, biovar eltor",
"parent": "A00"
},
{
"code": "A009",
"description": "Cholera, unspecified",
"parent": "A00"
}
],
"lastPage": true
}Get an entity from ICD10 terminology with a specific value/description/custom property.
Here we’re looking for an entity with a specific description value of Cholera:
GET https://sandbox.better.care/terminology-adapter/rest/terminology/codesystem/ICD10/entities/query?description=Cholera&match=exact
[
{
"code": "A00",
"description": "Cholera"
}
]Get children of a specific entity, in our case an entity with the code A00:
GET https://sandbox.better.care/terminology-adapter/rest/terminology/codesystem/ICD10/entity/A00/children
[
{
"code": "A000",
"description": "Cholera due to Vibrio cholerae 01, biovar cholerae",
"parent": "A00"
},
{
"code": "A001",
"description": "Cholera due to Vibrio cholerae 01, biovar eltor",
"parent": "A00"
},
{
"code": "A009",
"description": "Cholera, unspecified",
"parent": "A00"
}
]Resource Store API
OpenEHR repository is not meant to store large binary resources. The Reference model type DV_MULTIMEDIA can include byte streams, but the recommendation is to contain no more than a small thumbnail, a representation of a larger resource.
Usually you store binary resources in a dedicated object storage solution of your choice, that returns an uniquely URI for the resource. That URI then becomes a part of the composition stored in the Platform.
To speed up the development, Better Platform includes a Resource Store service, which accepts binary resources and can serve them when needed.
|
Resource Store service is not meant to be used in production environment. It implements only basic security mechanisms which are vulnerable to brute-force attack and could expose sensitive data. |
Store a binary resource
You can store a single resource as a binary stream or as a multipart/form-data encodined request.
Binary stream
| parameter | type | required | description |
|---|---|---|---|
mimeType |
string |
yes |
Has to be one of the IANA media types, describing the type of the resource. |
fileName |
string |
no |
Pass the resource filename. |
The request body should contain the resource encoded as a application/octet-stream.
To upload an image, set the mimeType correctly:
POST https://sandbox.better.care/store/rest?mimeType=image/png&fileName=image.png
{
"mimeType": "image/png",
"href": "https://sandbox.better.care/store/rest/06fc74ea-998b-4978-ab49-e69900bfff0e",
"fileName": "image.png",
"size": 165396
}Multipart form-data
No parameters are required.
The request body should contain the resource encoded as a multipart/form-data.
POST https://sandbox.better.care/store/rest/form
{
"mimeType": "image/png",
"href": "https://sandbox.better.care/store/rest/06fc74ea-998b-4978-ab49-e69900bfff0e",
"fileName": "image.png",
"size": 165396
}Get resource metadata
Get resource info with this call.
Pass the resourceId as the path element in:
GET https://sandbox.better.care/store/rest/<resourceId>/meta
GET https://sandbox.better.care/store/rest/06fc74ea-998b-4978-ab49-e69900bfff0e/meta
{
"mimeType": "image/png",
"href": "https://sandbox.better.care/store/rest/06fc74ea-998b-4978-ab49-e69900bfff0e",
"fileName": "image.png",
"size": 165396
}Clinical Decision Support API
Healthcare domain often includes clinical logic, various calculations when assessing risks, categorizing different patient related aspects or prescribing or dispensing medications.
Such actions are precise, sometimes need a lot of input data and or data transformations. Implementing these operations is very error prone and needs to be thoroughly tested.
In order to overcome such difficulties, openEHR introduced Clinical Decision Guides as a sort of instrument to provide a mean to define these actions in a uniform way.
Guides
A guide is a script, written in a notation called GDL (guide definition language). It usually contains a lot of if-then expressions, it can query data from the openEHR repository and produce results, which help driving at-point-of-care decision support applications.
Better Platform offers guide storage, retrieval and execution environment.
| Better Platform does not include tools to author such documents. |
List guides
Get a list of all guides stored in Better Platform.
GET https://sandbox.better.care/thinkcds/rest/v1/guide
[
{
"id": "Insufficient diabetes control",
"name": "Insufficient diabetes control",
"description": "Evaluates patients with diagnosed diabetes and categorises them into high risk groups. 1",
"use": "Alert doctor when treating a patient with insufficient diabetes control.",
"persistResult": false
},
{
"id": "delays trombolitic therapy",
"name": "AF and Acute Coronary Syndrome or ATC stent",
"description": "Antithrombotic therapy for AF and Acute Coronary Syndrome or ATC/stent",
"purpose": "Indication for antithrombotic therapy in patients with AF and Acute Coronary Syndrome or ATC/stent",
"use": "Choose antithrombotic therapy in patients with AF and Acute Coronary Syndrome or ATC/stent",
"references": "European Heart Journal, 2012 focussed update of the ESC Guidelines for the management of atrial fibrillation",
"persistResult": false
},
{
"id": "AF treatment_duration",
"name": "Guideline for AF treatment",
"description": "Medication for prevention thromboembolism in AF patient",
"purpose": "Verify medication for AF treatment, pre and post cardioversion",
"misuse": "",
"use": "Nonvalvular atrial fibrillation",
"persistResult": false
},
{
"id": "das28",
"name": "DAS28",
"description": "Calculates DAS28 score from observation data.",
"purpose": "A GDL tool to assess Rheumatoid Arthritis disease activity.",
"misuse": "Not for use outside of DAS28 or licensing terms.",
"use": "For use as part of rheumatology assessments.",
"references": "http://www.das-score.nl/das28/en/introduction-menu.html",
"persistResult": false
},
{
"id": "BMI_categorisation",
"name": "Body Mass Index calculation",
"use": "It is used to catgorise patients into 4 weight categories: Underweight Normal Overweight Obese",
"persistResult": false
}
]Upload a new guide
Once you have the guide of your interest in the GDL format, you can upload it to the internal Better Platform for storage and execution.
Find here a sample guide:
Our sample BMI categorisation guide will load the latest body weight and body height data from the repository and calculate the BMI value.
On base of the calculated value it will provide a categorisation value of either Underweight, Normal, Overweight or Obese.
Sample BMI guide is available here.
If you want to test it, make sure you upload it to the EHR Server.
In our examples the uploaded guide name is BMI_categorisation.
POST https://sandbox.better.care/thinkcds/rest/v1/guide`
Request body must contain the guide.
Any 2xx range response represents a successful action.
Delete a guide
Delete a guide by passing its ID in the request path:
DELETE https://sandbox.better.care/thinkcds/rest/v1/guide/<guideId>`
DELETE https://sandbox.better.care/thinkcds/rest/v1/guide/BMI_categorisation`
Any 2xx range response represents a successful action.
Execute a guide
Once you have a guide you would like to execute, it is up to you to provide all necessary inputs for it to execute smoothly.
Usually you need to provide:
-
provide access to the repository/EHR containing the data, or
-
input data in form of a composition containing all the data.
We will work with the following composition:
{
"vital_signs/_uid": "d099e9b3-5c4a-4a7f-9232-fc198ad357b0::andrazk-demo.sandbox.com::1",
"vital_signs/language|code": "en",
"vital_signs/language|terminology": "ISO_639-1",
"vital_signs/territory|code": "US",
"vital_signs/territory|terminology": "ISO_3166-1",
"vital_signs/context/start_time": "2019-05-16T15:12:33.16+02:00",
"vital_signs/context/setting|code": "238",
"vital_signs/context/setting|value": "other care",
"vital_signs/context/setting|terminology": "openehr",
"vital_signs/height_length:0/any_event:0/body_height_length|magnitude": 190,
"vital_signs/height_length:0/any_event:0/body_height_length|unit": "cm",
"vital_signs/height_length:0/any_event:0/time": "2019-05-16T15:12:33.16+02:00",
"vital_signs/height_length:0/language|code": "en",
"vital_signs/height_length:0/language|terminology": "ISO_639-1",
"vital_signs/height_length:0/encoding|code": "UTF-8",
"vital_signs/height_length:0/encoding|terminology": "IANA_character-sets",
"vital_signs/body_weight:0/any_event:0/body_weight|magnitude": 89,
"vital_signs/body_weight:0/any_event:0/body_weight|unit": "kg",
"vital_signs/body_weight:0/any_event:0/time": "2019-05-16T15:12:33.16+02:00",
"vital_signs/body_weight:0/language|code": "en",
"vital_signs/body_weight:0/language|terminology": "ISO_639-1",
"vital_signs/body_weight:0/encoding|code": "UTF-8",
"vital_signs/body_weight:0/encoding|terminology": "IANA_character-sets",
"vital_signs/category|code": "433",
"vital_signs/category|value": "event",
"vital_signs/category|terminology": "openehr",
"vital_signs/composer|name": "andrazk"
}As you see, it contains both height and weight data needed to calculate the BMI.
Execute a guide on specific EHR IDs
To execute a guide based on a query, you need to provide the guide ID, access to the data in the repository and a comma separated list of EHR IDs with which to work.
GET https://sandbox.better.care/thinkcds/rest/v1/guide/<guideId>/execute/<ehrUids>/query
For this example, we will assume the actual EHR contains the data needed to execute the sample guide.
| It is not a requirement that the data should be contained in a single composition. The guide will query the data according to its logic. |
| parameter | type | required | description |
|---|---|---|---|
Authorization |
string |
yes |
Query will access the Better Platform repository with an external request, hence it needs an authorization header for that request.
The username passed needs to have access to the |
format |
string |
no, |
This parameter can be:
|
persist |
boolean |
no, |
If set to |
trace |
boolean |
no, |
When set to |
templateId |
string |
no, |
Define the template which will be used to base the composition contents on. |
GET https://sandbox.better.care/thinkcds/rest/v1/guide/BMI_categorisation/execute/fd76b3a7-6813-430d-b6b7-e7e5e935b5f3/query?trace=true&persist=true&format=plain
[
{
"ehrId": "fd76b3a7-6813-430d-b6b7-e7e5e935b5f3",
"results": [
{
"name": "gt0002",
"value": {
"otherReferenceRanges": [],
"magnitude": 89,
"units": "kg"
},
"archetypeId": "openEHR-EHR-OBSERVATION.body_weight.v1",
"archetypeElementReference": "/data[at0002]/events[at0003]/data[at0001]/items[at0004]"
},
{
"name": "gt0003",
"value": {
"otherReferenceRanges": [],
"magnitude": 190,
"units": "cm"
},
"archetypeId": "openEHR-EHR-OBSERVATION.height.v1",
"archetypeElementReference": "/data[at0001]/events[at0002]/data[at0003]/items[at0004]"
},
{
"name": "gt0004",
"value": {
"otherReferenceRanges": [],
"magnitude": 24.65373961,
"units": "kg/m2",
"precision": 2
},
"archetypeId": "openEHR-EHR-OBSERVATION.body_mass_index.v1",
"archetypeElementReference": "/data[at0001]/events[at0002]/data[at0003]/items[at0004]"
},
{
"name": "gt0006",
"value": {
"value": "Normal",
"mappings": []
},
"archetypeId": "openEHR-EHR-EVALUATION.gdl_result_details.v1",
"archetypeElementReference": "/data[at0001]/items[at0015]"
}
],
"trace": {
"rules": [
{
"id": "gt0001",
"text": "Calculate body mass index",
"executed": true,
"inputs": [
{
"variableCode": "gt0002",
"variableName": "Weight",
"value": {
"otherReferenceRanges": [],
"magnitude": 89,
"units": "kg"
}
},
{
"variableCode": "gt0003",
"variableName": "Height/Length",
"value": {
"otherReferenceRanges": [],
"magnitude": 190,
"units": "cm"
}
}
],
"outputs": [
{
"type": "assign",
"variableCode": "gt0004",
"variableText": "Body Mass Index",
"variableAttribute": "precision",
"newValue": 2
},
{
"type": "assign",
"variableCode": "gt0004",
"variableText": "Body Mass Index",
"variableAttribute": "units",
"newValue": "kg/m2"
},
{
"type": "assign",
"variableCode": "gt0004",
"variableText": "Body Mass Index",
"variableAttribute": "magnitude",
"newValue": 24.65373961
}
]
},
{
"id": "gt0005",
"text": "Set Underweight",
"executed": false,
"inputs": [
{
"variableCode": "gt0004",
"variableName": "Body Mass Index",
"value": {
"otherReferenceRanges": [],
"magnitude": 24.65373961,
"units": "kg/m2",
"precision": 2
}
}
],
"outputs": []
},
{
"id": "gt0007",
"text": "Set Normal",
"executed": true,
"inputs": [
{
"variableCode": "gt0004",
"variableName": "Body Mass Index",
"value": {
"otherReferenceRanges": [],
"magnitude": 24.65373961,
"units": "kg/m2",
"precision": 2
}
}
],
"outputs": [
{
"type": "assign",
"variableCode": "gt0006",
"variableText": "Result",
"newValue": {
"value": "Normal",
"mappings": []
}
}
]
},
{
"id": "gt0008",
"text": "Set Overweight",
"executed": false,
"inputs": [
{
"variableCode": "gt0004",
"variableName": "Body Mass Index",
"value": {
"otherReferenceRanges": [],
"magnitude": 24.65373961,
"units": "kg/m2",
"precision": 2
}
}
],
"outputs": []
},
{
"id": "gt0009",
"text": "Set Obese",
"executed": false,
"inputs": [
{
"variableCode": "gt0004",
"variableName": "Body Mass Index",
"value": {
"otherReferenceRanges": [],
"magnitude": 24.65373961,
"units": "kg/m2",
"precision": 2
}
}
],
"outputs": []
}
],
"storedCompositionId": "ddb03bdf-75eb-442d-b8c3-8efc3c1edb44::andrazk-demo.sandbox.com::1"
}
}
]Execute guide on a specific composition
Similar to executing a guide against a repository, you can execute a guide over a single composition, which should contain all the necessary data, instead of running the query on top of the repository
Template ID is now a required request path parameter.
POST https://sandbox.better.care/thinkcds/rest/v1/guide/<guideId>/execute/<ehrIds>/composition/<templateId>
POST https://sandbox.better.care/thinkcds/rest/v1/guide/BMI_categorisation/execute/fd76b3a7-6813-430d-b6b7-e7e5e935b5f3/composition/Vital%20Signs
Request body must contain the composition in either FLAT or STRUCTURED format.
Please see executing a guide with a query.
OpenEHR API
Why it exists?
OpenEHR format is open, vendor-neutral data format. Once data is stored in such an archetype based format, we reach high levels of interoperability. Any solution operating on top of OpenEHR based CDR can use data from any other OpenEHR based CDR.
To facilitate the access to such data, OpenEHR Foundation has also specified an OpenEHR REST API interface, which provides a unified access to basic services in order to get to the data:
-
EHR management,
-
composition handling,
-
data querying.
All OpenEHR based Server solutions that implement this API are fully interchangeable on a basic level of services.
Specification
Specification in Apiary format can be found here:
https://specifications.openehr.org/releases/ITS-REST/latest/
It is up to the OpenEHR Server provider to decide whether to implement this API or not.
While OpenEHR REST API provides basic services and data access, efficient solutions might require other services, implemented by the OpenEHR Server provider, to facilitate and speed up the development.